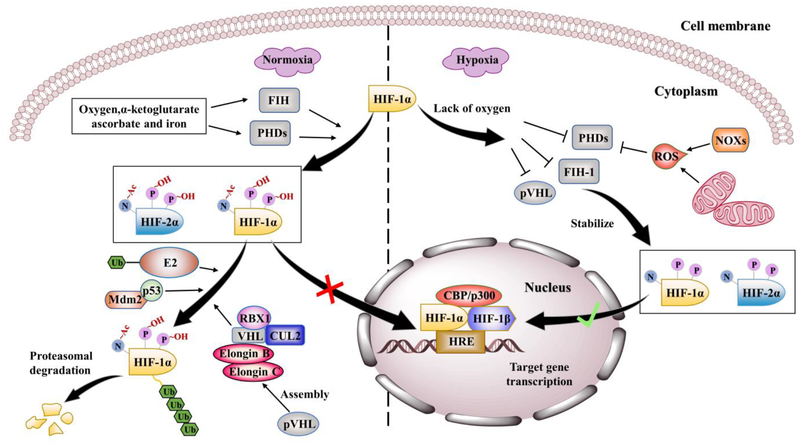
Figure 2.
Schematic diagram of canonical mechanisms regulating HIF-1/2. Under normoxic conditions (left panel), PHDs and FIH hydroxylate HIF-1/2α on proline residues and asparagine residue and trigger formation of hydroxylated HIF-1/2α. In the meantime, pVHL mediates the assembly of a complex containing VHL, Elongin B, Elongin C, CUL2 and RBX1, which binds ubiquitin-conjugated E2 component to accomplish ubiquitination of HIF-1/2α proteins. P53 is able to recruit an E3 ubiquitin–protein ligase, Mdm2, to help the proteasomal degradation of HIF-1α mediated by an E2 and E3 ubiquitin ligase–pVHL complex. Besides, hydroxylation of the asparagine residue in the C-TAD of HIF-1/2, FIH blocks the essential interaction between HIF-1α and co-activators such as CBP/p300. However, under hypoxic conditions (right panel), pVHL, PHDs and FIH activities are inhibited by limited oxygen, and ROS generation mechanisms in mitochondria and others such as NADPH oxidases (NOXs), leading to the escape of HIF-1/2α from proteasomal degradation. Thus, the HIF transcriptional complex binds to the HREs motif on the DNA of target genes and activates the target gene transcription.