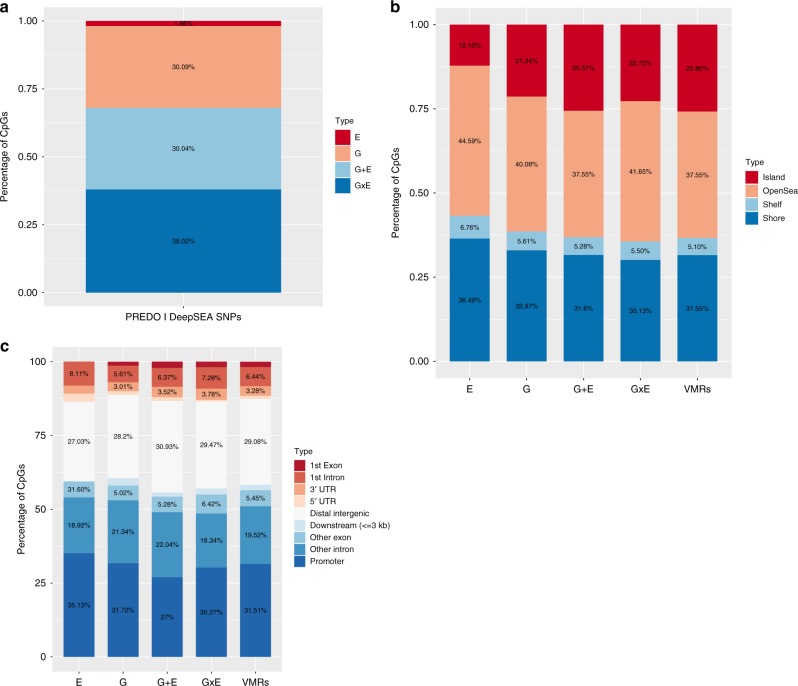
Fig. 3.
VMR analysis in DeepSEA annotated SNPs in PREDO I dataset. a Percentage of models (G, E, GxE or G + E) with the lowest AIC explaining variable DNA methylation using the PREDO I dataset with DeepSEA annotated SNPs. b Distribution of the locations of all VMRs and tagVMRs with best model E, G, G + E and GxE on the 450k array using only DeepSEA variants in relationship to CpG-Islands based on the Illumina 450 K annotation. c Distribution of gene-centric locations of all VMRs and tagVMRs with best model E, G, G + E and GxE on the 450k array using only DeepSEA variants