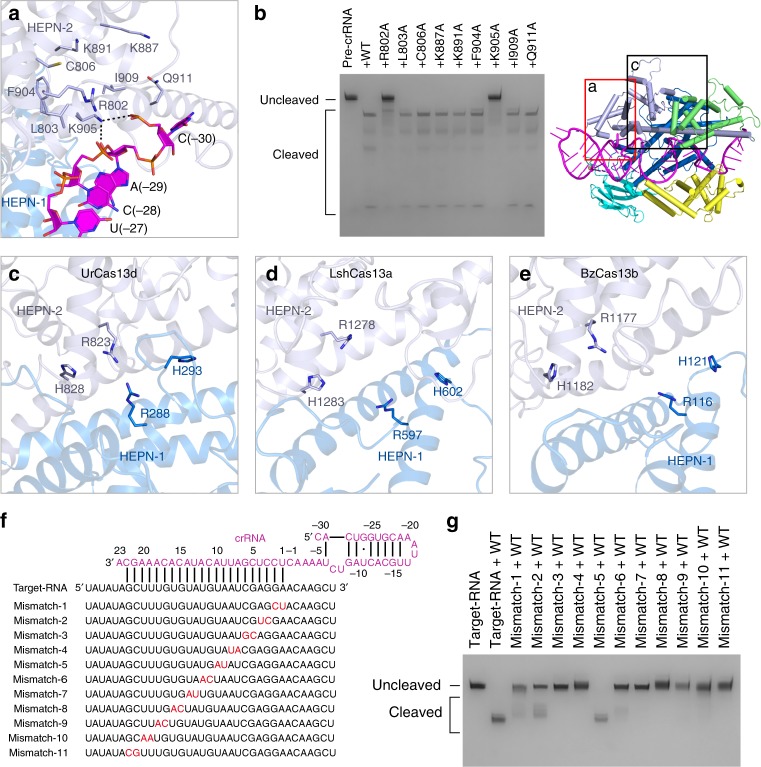
Fig. 6.
Active sites involved in pre-crRNA processing and target cleavage. a Close-up view of UrCas13d active site for pre-crRNA processing. Hydrogen bonds are shown as dashed lines. Domains and residues are colored according to Fig. 1. b Denaturing gel shows pre-crRNA cleavage by wild-type UrCas13d and the mutants speculatively involved in pre-crRNA processing. c Close-up view of UrCas13d active site for target RNA cleavage (PDB code: 6IV9). Residues A288 and A823 of the UrCas13d R288A/R823A binary complex was virtually mutated back to R288 and R823. d Close-up view of LshCas13a (Leptotrichia shahii Cas13a) active site for target RNA cleavage (PDB code: 5WTK). e Close-up view of BzCas13b (Bergeyella zoohelcum Cas13b) active site for target RNA cleavage. Residue A1177 of the BzCas13b R1177A binary complex (PDB code: 6AAY) was virtually mutated back to R1177. f Schematic representations of crRNA, target RNA and the mismatched targets designed for testing the mismatch tolerance between the crRNA spacer region and target RNA in UrCas13d. g Denaturing gel demonstrating the cleavage of target RNA and the mismatched target RNAs by wild-type UrCas13d in complex with crRNA