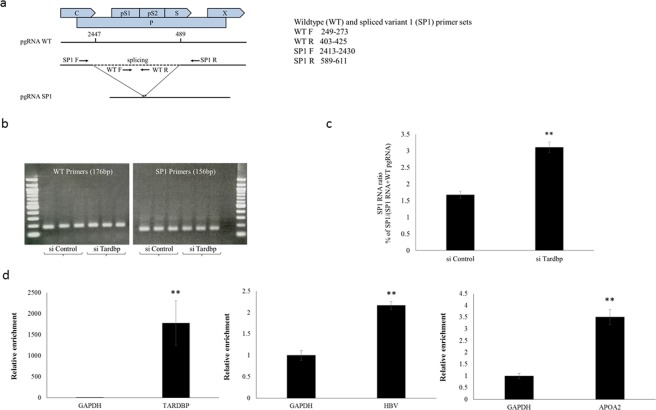
Figure 8.
TARDBP regulates HBV pgRNA splicing. (a) A schematic representation of the HBV genome indicating the site of splicing for the major splice variant, SP1 (intron 2447/489), and the primers designed for PCR amplification of the wild type (WT) and SP1 pgRNA. (b) The T23 cells, which stably express HBV, were transfected in triplicate with siRNAs against TARDBP or control. Total RNA was extracted from the cells, followed by reverse transcription. The resulting cDNAs were analyzed by PCR followed by agarose gel electrophoresis with primers specific for WT pgRNA or SP1 RNA. The results confirmed that there was no non-specific amplification of WT products with SP1 primers and that the two products were at the expected sizes. (c) The primers in (b) were henceforth utilized in a quantitative PCR of the samples to determine the proportion of SP1 RNA, which was calculated as SP1 RNA/(SP1 pgRNA + WT pgRNA). (d) Whole cell lysate from FLAG-TARDBP or FLAG-transfected T23 cells was immunoprecipitated with an anti-FLAG antibody and subjected to isolation of RNA and qPCR analysis using primers for TARDBP, APOA2 and total HBV mRNAs. The amount of mRNA that was precipitated from the TARDBP transfected cells was calculated as a ratio relative to that bound to the control-transfected cells for each of the primers. Relative mRNA values are presented as a ratio with respect to the amount of GAPDH mRNA, which was set at one. Results are reported as mean ± SD, while **P < 0.01.