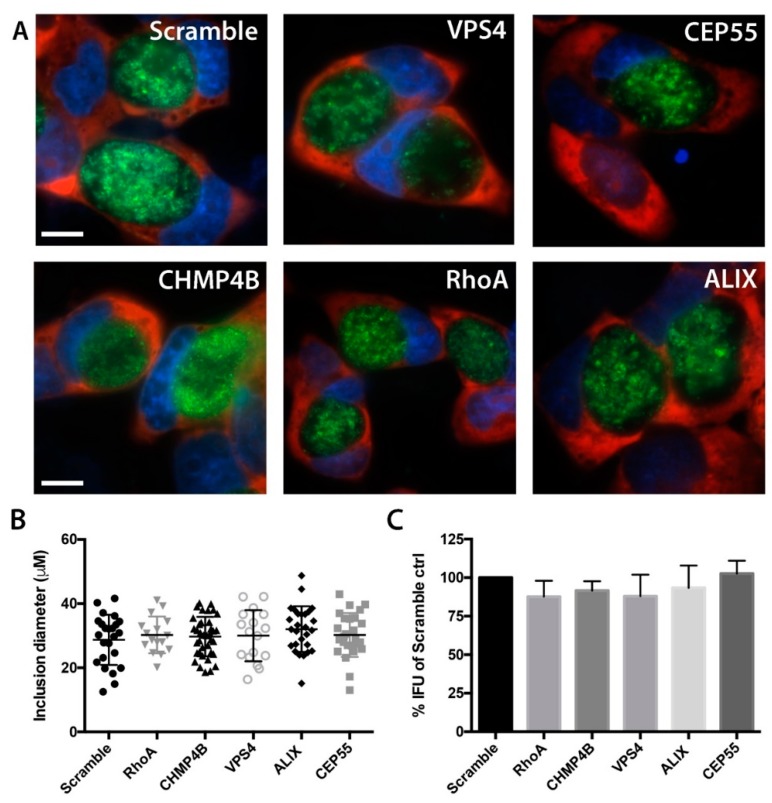
Figure 4.
Primary C. trachomatis infections are not affected by abscission protein RNAi. (A) Morphologies of C. trachomatis inclusions at 48 hpi following siRNA treatments with a nontargeting control (Scramble), VPS4, CEP55, CHMP4B, RhoA, and ALIX. Images were acquired at 60X magnification. GFP-expressing C. trachomatis (green), mCherry-HeLa cells (red), and DAPI-labeled nuclei (blue) are labeled. Scale bar = 10 μm. (B) Inclusion diameters of all inclusions in 10 images per host target knockdown. Diameter measurements were performed from nucleus side of the inclusion outward. (C) IFU analysis of Chlamydia-infected HeLa cells following knockdown with abscission targets. For graphs in B and C, n = 3, and no significance was measured using one way ANOVA with Dunnett’s multiple comparisons test in inclusion diameter or IFU compared to Scramble control.