Abstract
Background:
Genome-wide association studies (GWAS) have begun to identify loci related to alcohol consumption, but little is known about whether this genetic propensity overlaps with specific indices of problem drinking in ascertained samples.
Methods:
In 6,731 European-Americans who had been exposed to alcohol, we examined whether polygenic risk scores (PRS) from a GWAS of weekly alcohol consumption in the UK Biobank predicted variance in 6 alcohol-related phenotypes: alcohol use, maximum drinks within 24 hours (MAXD), total score on the self-rating of effects of ethanol questionnaire (SRE-T), DSM-IV alcohol dependence (DSM4AD), DSM-5 alcohol use disorder symptom counts (DSM5AUDSX), and reduction/cessation of problematic drinking. We also examined the extent to which an SNP (rs1229984) in ADH1B, which is strongly associated with both alcohol consumption and dependence, contributed to the polygenic association with these phenotypes and whether PRS interacted with sex, age, or family history of alcoholism to predict alcohol-related outcomes. We performed mixed effect regression analyses, with family membership and recruitment site included as random effects, as well as survival modeling of age of onset of DSM-IV alcohol dependence.
Results:
PRS for alcohol consumption significantly predicted variance in five of the six outcomes: alcohol use (Δmarginal R2 =1.39%, ΔAUC=0.011), DSM4AD (Δmarginal R2=0.56%; ΔAUC=0.003), DSM5AUDSX (Δmarginal R2 =0.49%), MAXD (Δmarginal R2 = 0.31%), and SRE-T (Δmarginal R2 = 0.22%). PRS were also associated with onset of DSM4AD (hazard ratio = 1.11, p = 2.08e-5). The inclusion of rs1229984 attenuated the effects of the alcohol consumption PRS, particularly for DSM4AD and DSM5AUDSX, but the PRS continued to exert an independent effect for all five alcohol measures (Δmarginal R2 after controlling for ADH1B = 0.14 – 1.22%). Interactions between PRS and sex, age or family history were nonsignificant.
Conclusion:
Genetic propensity for typical alcohol consumption was associated with alcohol use, and was also associated with 4 of the additional 5 outcomes, though the variance explained in this sample was modest. Future GWAS that focus on the multi-faceted nature of alcohol use disorder, which goes beyond consumption, might reveal additional information regarding the polygenic underpinnings of problem drinking.
Keywords: alcohol dependence, alcohol consumption, polygenic risk, ADH1B
INTRODUCTION
Alcohol consumption ranges from infrequent intake of alcoholic beverages, through more frequent and heavy episodic drinking (e.g., bingeing), to drinking at problematic levels that are potentially indicative of alcohol use disorders (AUD). While there is considerable debate surrounding the putative benefits of moderate drinking (Stockwell et al., 2016; Wood et al., 2018), excessive alcohol consumption has been unequivocally identified as one of the top 10 contributors to worldwide morbidity and mortality (World Health Organization, 2014). Excessive alcohol consumption can escalate to AUD diagnosis when accompanied by other psychological, behavioral, and physical symptoms, such as loss of control over drinking, tolerance, withdrawal and persistent drinking despite health problems (Wise & Koob, 2014). It is estimated that AUD attributable costs approximate 1% of the gross domestic product of developed nations (Rehm et al., 2009).
Both alcohol consumption and AUD (as well as other indices of problematic drinking, e.g., the problem subscale of the Alcohol Use Disorders Identification Test (AUDIT-P) (Sanchez-Roige et al., 2017)) are heritable. Twin studies suggest that additive genetic factors contribute to 40–70% of the variance in alcohol consumption and AUD (Heath & Martin, 1994; Prescott & Kendler, 1999; Verhulst et al., 2015). The degree to which there is overlap in the genetic factors contributing to alcohol consumption vs. AUD is less clear from twin data (Agrawal, Lynskey, Heath, & Chassin, 2011; Dick, Meyers, Rose, Kaprio, & Kendler, 2011; Grant et al., 2009; Kenneth S Kendler, Myers, Dick, & Prescott, 2010), but recent genome-wide association studies (GWAS) have estimated this overlap using Linkage Disequilibrium Score Regression (LDSC (Bulik-Sullivan et al., 2015)). Based on GWAS of both alcohol consumption (N=112, 117) (Clarke et al., 2017) and DSM-IV alcohol dependence (N=14,904 cases and 37,944 controls) (Walters et al., 2018), common variants explain, in aggregate, between 9–13% of the variance in these phenotypes (i.e., SNP heritability) and the estimated genetic correlation between them ranges from rg = 0.37 – 0.70 (Walters et al., 2018).
As the correlation between consumption measures and dependence is derived from composite phenotypes (e.g., alcohol dependence diagnosis using structured interviews or clinician reports, consumption via self-reports) and in heterogeneous samples, further work is needed to pinpoint which specific aspects of drinking (e.g. lifetime alcohol use vs. alcohol dependence diagnosis) are most genetically correlated with self-reported levels of alcohol consumption. To study particular features of problem drinking, ascertained samples that are enriched for liability to alcohol dependence may provide increased power through larger sample sizes as well as detailed phenotyping. However, it is likely that several factors might moderate the extent to which genetic liability for alcohol consumption in a specific discovery population, such as the older volunteer cohort that comprises the UK Biobank, is associated with problem drinking in an independent sample. One possibility is that genetic overlap might be more pronounced in a similar age group as that comprising the discovery GWAS, possibly due to cohort-specific genetic and environmental influences. Other moderators might include sex, cultural effects, and access to alcohol, as well as family history of alcohol problems. For instance, family history has been shown in other studies to accentuate the impact of genetic vulnerability on psychiatrically relevant outcomes (e.g., Agerbo et al., 2015).
Beyond genetic correlations from LDSC, polygenic risk scores (PRS) offer an alternative approach for demonstrating genetic overlap between two traits (e.g. alcohol consumption and AUD). PRS represent the additive effects of independent single nucleotide polymorphisms (SNPs) that are weighted by their effect sizes from a “discovery” GWAS (International Schizophrenia Consortium, 2009). With this approach, every individual in the independent “target” sample is assigned a score that indexes their estimated genetic propensity to the behavior studied in the discovery GWAS. The phenotype of interest in the target sample is then regressed on the polygenic score, and the strength of this association is assessed using R2 or other measures of predictor efficacy (e.g., Area Under the Curve (AUC)). Although the PRS incorporates additional SNPs beyond those meeting the stringent genome-wide significance threshold, PRS typically explain a very small percentage (usually <10%) of the variance in the target sample phenotype (Dudbridge, 2013; Wray et al., 2014). How much variance PRS explain is dependent upon the SNP heritability of the phenotype, the size of both the discovery GWAS and the target sample, the selection thresholds for SNPs included in the PRS, and the methods used for weighting the effect sizes (Dudbridge, 2013). The extent to which a PRS derived from a GWAS of alcohol consumption is associated with aspects of problem drinking is, therefore, an estimate of their genetic commonality, although causal processes can also be represented (Swerdlow et al., 2016).
To date, the most robust genetic signals identified for both alcohol consumption and dependence have been within ADH1B (across multiple ancestries (Bierut et al., 2012; Clarke et al., 2017; Kranzler et al., 2019; Luczak et al., 2006; Sanchez-Roige et al., 2018; Walters et al., 2018). Recent GWAS of alcohol consumption (Jorgenson et al., 2017; Liu et al., 2019; Schumann et al., 2011, 2016) have implicated loci in AUTS2, KLB, GCKR and other genes; however, evidence for their involvement in the genetics of alcohol dependence remains limited (Sanchez-Roige et al., 2018), although a recent preprint implicated GCKR in a large GWAS of AUD (Kranzler et al., 2019). The protective allele of rs1229984, the missense SNP within ADH1B that affects the conversion of ethanol to acetaldehyde, exerts one of the largest single-variant effects on a polygenic trait, with up to a 3-fold decrease in risk for alcohol dependence (Edenberg, 2007; Edenberg & McClintick, 2018). Thus, studies that examine the polygenic overlap between consumption and dependence should account for the role of ADH1B in aggregated genomic propensity.
In this study, we use a large (N = 6,731), deeply-phenotyped target sample of European-Americans who reported ever drinking alcohol (and enriched for those with alcohol dependence) and a very large (N = 112,117) discovery GWAS in a European-only volunteer cohort (the UK Biobank) to examine the genetic overlap between alcohol consumption and non-problem as well as problematic drinking behaviors. Our target sample consists of individuals from the Collaborative Study of the Genetics of Alcoholism (COGA) who have been assessed one or more times using an instrument specifically designed to evaluate risk for substance use and common psychiatric disorders. We utilized a PRS approach to examine the polygenic overlap between self-reported alcohol consumption in the UK Biobank volunteer cohort and several aspects of drinking in COGA. We aimed to leverage the unique strengths of both samples in this study to ask (a) whether a polygenic score for alcohol consumption (average intake per week) from a population-based cohort predicts a range of drinking milestones in a sample that is enriched for familial risk for alcohol problems, (b) whether any association between the PRS and drinking milestones is moderated by age, sex, or family history of alcohol use disorders, and (c) whether the PRS is associated with aspects of drinking above and beyond the effect of the strongest signal for AUD, rs1229984 in ADH1B.
METHODS
Sample:
The Collaborative Study on the Genetics of Alcoholism (COGA) currently consists of 12,145 individuals with GWAS data. The goal of COGA is to elucidate the genetic underpinnings of alcohol use disorders and problem drinking across the lifespan. The study is described in detail elsewhere (Begleiter et al., 1995; Bucholz et al., 2017). Briefly, probands were identified through primarily inpatient alcohol treatment programs at seven U.S. sites. Probands and their family members were invited to participate if they had a sufficiently large family (usually > 3 sibs with parents available) with two or more members in the COGA catchment area. Control families (2 parents and 3 or more offspring over the age of 14) were also selected from a variety of sources, (e.g., dental clinics, driver license registries). The Institutional Review Boards at all sites approved this study and written consent was obtained from all participants. As the PRS were derived from a sample of Europeans, only individuals identified as part of the European American subsample of COGA, determined using genomic data, were included in the analyses reported here (N = 7,645). To avoid confounding by individuals at high genetic risk who elected not to drink due to personal (e.g., religious, cultural, health) reasons, we also excluded individuals reporting no lifetime use of alcohol (N = 336). We also excluded those aged 12–19 (N = 578) as they may not be past the period of maximal risk for onset of alcohol use and problems, yielding a final analysis N = 6,731.
Genotyping:
Genotyping for the COGA European American participants was performed using the Illumina 1M, Illumina OmniExpress, and Illumina 2.5M (Illumina, San Diego, CA), and Smokescreen (BioRelm, Walnut, CA) arrays. Array type was included as a covariate in all analyses. A pruned set of 47,000 variants that were genotyped on all platforms and had minor allele frequencies (MAF) > 10% in the combined samples, Hardy-Weinberg Equilibrium (HWE) p-values > 0.001, missing rates < 2%, and were not in linkage disequilibrium (LD, defined as R2 < 0.5) were used to assess reported pedigree structure using identity-by-descent calculations in PLINK (Purcell et al., 2007). Family structures were altered as needed and SNP genotypes were tested for Mendelian inconsistencies with the revised family structure (O’Connell & Weeks, 1998). Genotype inconsistencies were set to missing. Imputation was to 1000 Genomes (EUR and AFR, Phase 3, b37, October 2014; build hg19) using SHAPEIT2 (Delaneau et al., 2012) and then Minimac3 (Das et al., 2016). Only non-palindromic variants, with missing rates < 5%, MAF > 3%, and HWE p values > 0.0001 were used for imputation. Imputed SNPs with information (INFO) scores < 0.30 or individual genotype probability scores < 0.90 were excluded. For the final dataset for PRS construction, palindromic SNPs (A/T or C/G), monomorphic SNPs, SNPs that did not pass Hardy-Weinberg equilibrium (HWE p < 1e-6), and SNPs with a minor allele frequency (MAF) less than 0.005 were excluded. In total, 6,881,872 SNPs passed quality control and data cleaning thresholds and were available for analysis. Genotypes that did not pass quality control prior to and upon imputation were excluded from PRS construction.
Measures:
All COGA participants were interviewed at least once using a version of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) ( Bucholz et al., 1994; Hesselbrock et al., 1999). For these analyses, an individual’s interview with the highest lifetime report for each measure was used. Measures included:
Alcohol use:
Drinking at least one drink a month for 6 or more consecutive months at any point in the lifetime (yes/no).
Maximum drinks (MAXD):
the maximum number of drinks ever consumed in a single 24-hour period (range = 1 – 100; values >100 (N = 32) were constrained to 100). To correct for skew, this variable was transformed by taking the natural logarithm of (MAXD).
Total score for the self-rating of the effects of ethanol (SRE-T):
The SRE-T score was derived from a self-report instrument administered to a smaller subset of COGA participants (N=4,296) and assesses the participant’s response to 4 items that address the number of drinks required for the participant to feel the effects of alcohol, feel dizzy or begin to slur in speech, to stumble, or to fall asleep. Scores across three time points (first five times the participant used alcohol, last three months and period of heaviest drinking) are averaged to create the total (SRE-T) score (Schuckit et al., 1997), winsorized at ± 2 standard deviations from the mean, and further transformed using the square root of the value to correct for skew.
DSM-IV Alcohol dependence (DSM4AD):
We elected to use DSM-IV (American Psychiatric Association, 2000) lifetime criteria for alcohol dependence, the endorsement of 3 or more of 7 dependence criteria that clustered within a single 12-month period, because it represents a more severe form of the diagnosis than the DSM-5. Age of onset of alcohol dependence (DSM-IV) was used in survival analyses, with those who had not met criteria for DSM4AD censored at their age at the last interview.
DSM-5 Alcohol use disorder symptom count (DSM5AUDSX):
We elected to use DSM-5 criteria for the quantitative index of AUD severity as it captures additional aspects of the disorder beyond DSM-IV dependence, thus providing a broader scale of liability. The lifetime endorsement of the 11 DSM-5 (American Psychiatric Association, 2013) criteria for AUD, summed at the interview with maximum number of symptoms, was used.
Reduction/cessation of problematic drinking:
An unordered categorical measure was created to represent three groups; (A) those who did not meet criteria for DSM5 AUD during their lifetime; (B) individuals who had a lifetime history of DSM-5 AUD and were current problematic drinkers due to an active AUD diagnosis in the past 12 months or were high-risk drinkers (defined as men: >= 5 drinks/day or >= 15 drinks in one week; women: >= 4 drinks/day or >= 8 in one week (NIAAA, 2004); and (C) those who had a lifetime history of AUD but had reduced/ceased their drinking and either did not report any AUD criteria (except craving), or were not high-risk drinkers, or were abstinent from alcohol, all in the past 12 months (McCutcheon et al., 2017; Schuckit et al., 2018). A comparison of Group A against either Group B or C contrasts presence or absence of a lifetime diagnosis of DSM-5 AUD, while the comparison of Group B and Group C stratifies those with a lifetime diagnosis into high risk drinkers, including those with active AUD, and low risk drinkers who may also be in abstinent remission.
Covariates included sex, participant’s age at their last interview, birth cohort (dummy variables representing birth years prior to 1930, 1930–1949, 1950–1969, and 1970 and after), the first three ancestral principal components, array type, recruitment site, and the total number of interviews with the participant.
Family history:
A binary measure representing whether at least one of the biological parents of the respondent had a history of DSM-5 AUD was used. Various sources of information were used to derive family history (e.g., parental interview, respondent report; see (Bucholz et al., 2017; McCutcheon et al., 2017) for details). Of the analytic sample, 1,656 individuals did not have a report on family history, and 485 individuals had a missing report on one parent while the other parent was confirmed to be family history negative. Thus, family history was not included as a covariate in all analyses and for analyses including family history (e.g., PRS*family history), the sample size was smaller (N = 5,075).
Statistical Analysis:
Construction of polygenic risk scores (PRS):
Effect sizes and effect alleles were derived from a GWAS of N = 112,117 unrelated European ancestry individuals in the UK Biobank (Clarke et al., 2017). Participants were asked about their current drinking status (never, previous, current, prefer not to say) and their average weekly and monthly consumption of a variety of alcoholic beverages (e.g. red wine, white wine, beer, spirits). An overall measure of average alcohol intake per week was derived from these measures. Age and weight were then regressed onto alcohol consumption in units per week in males and females separately, and the residuals from these regressions were then pooled (males + females) to form the alcohol consumption phenotype of interest. A GWAS was conducted with 12,489,782 quality-controlled SNPs and UKB assessment center, genotyping batch and 15 principal components included as additional covariates. In COGA, after removing palindromic/ambiguous SNPs from the summary statistics, PRS were coded for every individual by multiplying an individual’s number of effect alleles at a particular SNP by that SNP’s effect size (beta) from the discovery GWAS, then averaging across SNPs to create one score per person. Clumping was done using the European subset of the 1000 Genomes Phase 3 sample (Consortium, 2015) as an external linkage disequilibrium (LD) reference panel, using a 500 kb physical distance and an LD threshold of r2>=0.25. Scores representing effect sizes with increasingly lenient thresholds of statistical significance in the discovery GWAS were constructed (pT <0.0001, pT <0.001, pT <0.01, pT <0.05, pT <0.10, pT <0.20, pT <0.30, pT <0.40, pT <0.50). Scores were standardized before statistical analysis.
Data Analysis:
All analyses were conducted in R (R Core Team, 2017). First, mixed effect regressions were used to examine whether alcohol consumption PRS predicted (a) alcohol use, (b) MAXD, (c) DSM5AUDSX, (d) DSM4AD, (e) SRE-T, and (f) cessation/reduction of problem drinking, and to determine which PRS threshold (i.e., pT) was most predictive of the drinking measure based on the p-value and R2. For the cessation/reduction measure, groups were contrasted with each other using binary comparisons (e.g., Group B vs. Group C). All regressions controlled for the covariates mentioned above as fixed effects, while the family identifier and recruitment site were included in the models as random effects (family nested within site). To assess model fit and the relative amount of variance explained by the PRS, we used the ‘MuMIn’ package in R to calculate both marginal and conditional R2 for each mixed model (Barton, 2011; Nakagawa & Schielzeth, 2012). We use the marginal R2 to select the most predictive pT. The proportion of variance attributable to the PRS (Δmarginal R2) was estimated as the difference between the marginal R2 (i.e., Δmarginal) of the model that included covariates and the PRS and the model with covariates alone (i.e. marginal R2(full model) - marginal R2(model without PRS)). Further, Cox proportional hazards survival analyses for onset of DSM4AD were conducted using the survival and survminer packages in R (Kassambara, Kosinski, & Biecek, 2017; Therneau & Lumley, 2015) with the same covariates as above. Violations of the proportional hazards assumption for the PRS were tested using scaled Schoenfeld residuals. For graphical depiction of cumulative survival curves, quartiles of PRS were computed and hazard ratios were estimated across those categories, with adjustment for covariates. Family membership and recruitment site were included in the survival models using the “cluster()” function, which produces a robust estimate of standard errors that accounts for potential clustering on those two variables (Therneau & Grambsch, 2013; Therneau & Lumley, 2015).
Next, we selected the most predictive p-value threshold (pT) to test for PRS-by-age, PRS-by-sex and PRS-by-family history interactions. Each interaction was tested in an independent model with all other cross-term covariates included (Keller, 2014). Finally, since the strongest and most robustly validated genetic signal for alcohol consumption and dependence is rs1229984 in ADH1B, we examined the extent to which including rs1229984 genotype as a covariate attenuated the variance explained by the PRS, for traits where alcohol consumption PRS were significantly predictive. The potential attenuation was computed by contrasting a model that included covariates and rs1229984 (coded in the direction of the effect allele, i.e., those homozygous for the protective allele were coded as 2) with a model that included those terms as well as the PRS (i.e. Null model: Phenotype ~ rs1229984 + covariates vs. Full model: Phenotype ~ PRS + rs1229984 + covariates).
Two additional analytic considerations were made. First, there have been increasing concerns regarding the potential oversampling for susceptibility to smoking in a sub-component of the UK Biobank (the UK BiLEVE sample; see (Munafò et al., 2017)). To examine this possibility, alcohol consumption PRS were also regressed on a measure of maximum cigarettes smoked per day (however, any observed association might also reflect pleiotropy, given the genetic correlation between smoking and drinking). Second, even though pseudo-R2 is the most widely accepted index of predictive utility for PRS analyses of categorical outcomes, metrics such as Area Under the Curve (AUC) may provide a more conservative and precise estimate of their predictive capabilities for binary traits (Wray et al., 2013). Thus, we supplemented a subset of our analyses with estimates of AUC. Youden’s J index (sensitivity + specificity – 1) was used to generate the optimal cut-point in PRS for prediction of any significantly associated dichotomous traits (Youden, 1950).
The number of independent multiple tests across the six primary phenotypes (alcohol use, MAXD, SRE-T, DSM4AD, DSM5AUDSX, and reduction/cessation) was estimated by identifying the degree of phenotypic correlation across the measures of drinking using spectral decomposition of the data in matSpD (Nyholt, 2004). Five independent tests were estimated (Veff = 4.82). Thus, for the first set of analyses, where PRS (including 9 p-value thresholds) predicted measures of drinking, we corrected for 45 tests (9 PRS thresholds for each of 5 independent phenotypic tests), for a Bonferroni-corrected p value of 0.05/45 = 1.1e-03. For follow-up analyses (e.g., adjustment for rs1229984), where only a single PRS at one p-value threshold was used, a less stringent correction of 0.05/5=0.01 was implemented.
RESULTS
Characteristics of the COGA sample (N = 6,731) are provided in Table 1. As expected, nearly 90% of the sample reported a lifetime history of alcohol use (at least once a month for at least six consecutive months). The prevalence of problem drinking (including DSM4AD and DSM5AUD) was relatively high as well. For instance, the lifetime prevalence of DSM4AD was 33.8%, while 19.0%, 11.1%, and 22.5% endorsed 2–3, 4–5, and 6 or more DSM5AUD lifetime criteria, respectively.
Table 1:
Characteristics of 6,731 participants of European descent more than 20 years old in the Collaborative Study of the Genetics of Alcoholism (COGA).
| Overall (n=6,731) |
|
|---|---|
| Female (%) | |
| Born before 1930 | 17.6% |
| Born 1930–1949 | 41.7% |
| Born in 1950 – 1969 | 36.1% |
| Born in 1970 – now | 4.6% |
| Age at last interview (mean (SD)) | 38.697 (14.189) |
| Family history positivea (%) (N = 5,075) | 71.1% |
| Number of interviews (mean (SD)) | 2.246 (1.528) |
| Alcohol use | 89.9% |
| DSM-IV Alcohol dependence (DSM4AD) | 33.8% |
| Age at onset of alcohol dependence (mean, SD) | 33.3 (15.1) |
| Alcohol dependent with active AUD or high risk drinking | 63.9% |
| Reduction/Cessationb (N = 6,718) | |
| Active DSM-5 AUD or high riskc drinkers | 33.4% |
| Nonactive DSM-5 AUD who are currently abstinent or currently low risk drinkers | 16.0% |
| No lifetime DSM-5 AUD diagnosis – and low risk drinkers | 50.5% |
| Maximum drinks in 24hrs (MAXD: mean, SD) | 18.9 (16.4) |
| Self-rating of ethanol (SRE-T) score (N=4,337) | 2.21 (0.61) |
| DSM-5 Alcohol use disorder symptom count (DSM5AUDSX mean, SD) | 3.073.47) |
Family history positive: either or both parents have history of lifetime AUD
Reflects past-12 month report at final interview.
High risk defined as men: >= 5 drinks/day or >= 15 drinks in one week; women: >= 4 drinks/day or >= 8 in one week
As shown in Table 2, the alcohol consumption PRS predicted alcohol use, DSM4AD, and DSM5AUDSX at all pT, even after correction for multiple testing. It predicted the greatest variance in alcohol use (Δmarginal R2= 0.0139; PRS pT < 0.2), followed by DSM4AD (Δmarginal R2 = 0.0056; PRS pT < 0.01). DSM5AUDSX was also significantly associated with the PRS (Δmarginal R2= 0.0049; PRS pT < 0.5). MAXD was associated with the PRS at thresholds pt < 0.5 – 0.001 (maximum Δmarginal R2 = 0.0031), while SRE-T scores were only associated with the alcohol consumption PRS at pT < 0.1 (Δmarginal R2 = 0.0022). The results pattern for reduction/cessation of problem drinking suggests that the PRS were only associated with the difference between those with no lifetime DSM-5 AUD (group A) versus the group with active DSM-5 AUD and/or high-risk drinking (group B) [PRS pt < 0.10: Odds-Ratio, OR = 1.21, 95% C.I. = 1.14 – 1.27, p = 1.29e-8]. The PRS was not significantly associated with the comparisons between those with no lifetime DSM5AUD vs. lifetime DSM5AUD but now low risk/abstinent (group C); OR = 1.09, 95% CI = 1.01 – 1.18, p = 0.028) or between those with active DSM5AUD/high-risk and those who were low-risk/abstinent; OR = 0.94, 95% CI = 0.86 – 1.02, p = 0.105). Because of this limited association with the PRS, the reduction/cessation measure was not carried forward into the conditional (controlling for rs1229984) follow-up analysis. DSM4AD and DSM5AUDSX were strongly correlated with maximum cigarettes smoked (p < 0.001) but alcohol consumption PRS were not predictive of maximum cigarettes smoked (N = 5,436, p = 0.67).
Table 2: The percent variance for alcohol consumption and problem drinking indices in COGA explained by polygenic risk scores (PRS).
Percent variance explained was determined via ∆marginal R2. The most predictive threshold (based on p-value) for each outcome is bolded. Alcohol use was defined as having had at least 1 drink per month for at least 6 consecutive months. The SRE-T is the SRE score averaged across 3 time points (first five times the participant used alcohol, last three months and period of heaviest drinking). There were three pairwise comparisons for the reduction/cessation outcome: comparing (1) those with no lifetime AUD, (2) those with lifetime AUD and current symptoms or no symptoms but high-risk drinking, and (3) those who stopped drinking altogether (were abstinent for the past 12 months) or who were low risk drinking; only the comparison between those with no lifetime AUD and those with lifetime AUD and/or high-risk drinking is shown here.
| N SNPs | Alcohol Use | Maximum Drinks (MAXD) | Self-rating of effects of ethanol (SRE-T) | DSM-5 Alcohol use disorder symptom count (DSM5AUDSX) | DSM-IV alcohol dependence (DSM4AD) | Reduction/Cessation of problem drinking$ (No lifetime AUD vs. active AUD/high-risk drinking only) | |
|---|---|---|---|---|---|---|---|
| PT<0.5 | 313,399 | 1.11* | 0.26* | 0.18 | 0.49* | 0.57* | 0.69* |
| PT <0.4 | 268,741 | 1.13* | 0.25* | 0.18 | 0.47* | 0.52* | 0.61* |
| PT <0.3 | 217,799 | 1.28* | 0.25* | 0.18 | 0.46* | 0.47* | 0.67* |
| PT <0.2 | 159,282 | 1.39* | 0.26* | 0.17 | 0.48* | 0.49* | 0.75* |
| PT <0.1 | 91,805 | 1.27* | 0.31* | 0.22* | 0.44* | 0.59* | 0.83* |
| PT <0.05 | 51,951 | 1.35* | 0.27* | 0.18 | 0.36* | 0.48* | 0.72* |
| PT <0.01 | 13,626 | 1.04* | 0.29* | 0.16 | 0.37* | 0.56* | 0.45* |
| PT <0.001 | 2,048 | 0.48* | 0.18* | 0.17 | 0.22* | 0.36* | 0.32* |
| PT <0.0001 | 349 | 0.58* | 0.08 | 0.10 | 0.14* | 0.29* | 0.25 |
denotes significance at Bonferroni corrected p-value of 0.05/45 = 1.1e-3
computed using pairwise comparisons of logistic regression models; reported ∆R2 is for the comparison between those with no lifetime AUD vs. those with lifetime AUD and current symptoms or no symptoms but high-risk drinking; other comparisons (e.g. between those with active AUD and/or high-risk drinking vs. those with lifetime AUD who are currently abstinent or low-risk drinkers) were nonsignificant.
Even though PRS pT < 0.01 was associated with DSM4AD at α < 1.1e-3, the difference in AUC attributable to the addition of the PRS was modest (AUC-covariates = 0.710; AUC-covariates+PRS = 0.713; equality test χ2 = 6.62, p = 0.01). Results for alcohol use were far stronger: the AUC in a model where PRS alone predicted alcohol use was 0.57. Even in comparison to the model with covariates alone (AUC=0.688), addition of PRS resulted in a statistically significant increase in prediction of alcohol use (AUC = 0.70; equality test χ2 = 9.36, p = 0.0022). Youden’s J was 0.11 (sensitivity = 0.49, specificity = 0.62) and a PRS cut-point of 0.0524 was determined to maximize classification, but at a weak AUC of 0.56.
The correlations between the PRS and rs1229984 ranged from −0.15 to −0.05. As shown in Table 3, the variance predicted by the PRS was partially independent of the inclusion of rs1229984 in ADH1B. There was a significant association between rs1229984 genotype and alcohol use, DSM4AD, DSM5AUDSX, MAXD, and SRE-T with rs1229984 predicting 0.34 – 1.19% of the variance in these drinking measures. After including rs1229984 genotype as a covariate, the alcohol consumption PRS continued to predict 0.14 – 1.22% of the variance across the drinking measures. In particular, rs1229984 genotype had the least influence on alcohol use, for which the PRS predicted more variance than the ADH1B genotype (Δmarginal R2 for ADH1B conditional on PRS=0.49%; Δmarginal R2 for PRS conditional on ADH1B=1.22%).
Table 3:
Association between polygenic risk scores (PRS) derived for alcohol consumption and problem drinking indices, adjusting for rs1229984 (ADH1B).
| DSM-IV Alcohol Dependence (DSM4AD) | DSM-5 AUD Symptom Count (DSM5AUDSX) | Alcohol Use | SRE-T | Maximum Drinks (MAXD) | |
|---|---|---|---|---|---|
| Most predictive PRS | pT <0.01 | pT <0.5 | pT <0.2 | pT <0.1 | pT <0.1 |
| Δmarginal R2 for PRS | 0.562% | 0.490% | 1.389% | 0.217% | 0.308% |
| Δmarginal R2 for ADH1B | 1.19% | 0.527% | 0.549% | 0.403% | 0.347% |
| Beta (SE) for ADH1B | −0.951 (0.159) p = 2.25e-09 | −0.967 (0.165) p = 5.31e-09 | −0.674 (0.159) p = 2.15e-05 | −0.165 (0.035) p = 2.65e-06 | −0.204 (0.040) p = 3.10e-07 |
| Δmarginal R2 for ADH1B, adjusting for PRS | 1.13% | 0.486% | 0.489% | 0.380% | 0.319% |
| Δmarginal R2 for PRS, adjusting for ADH1B | 0.420% | 0.395% | 1.22% | 0.141% | 0.247% |
| Beta (SE) for PRS, after adjusting for ADH1B | 0.144 (0.032) p = 6.31e-06 | 0.221 (0.040) p = 2.65e-08 | 0.235 (0.046) p = 3.90e-07 | 0.023 (0.008) p = 0.006 | 0.045 (0.010) p = 1.81e-06 |
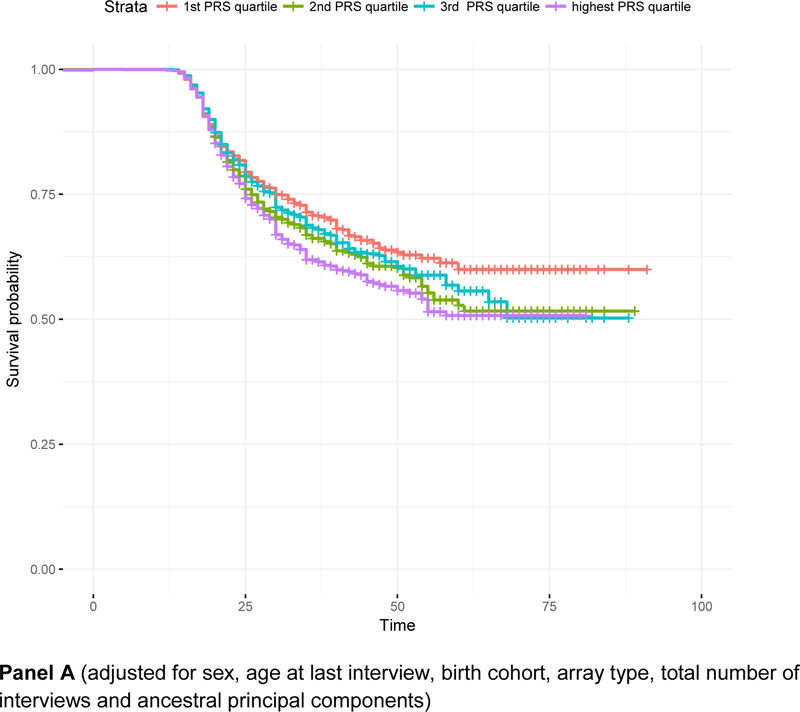
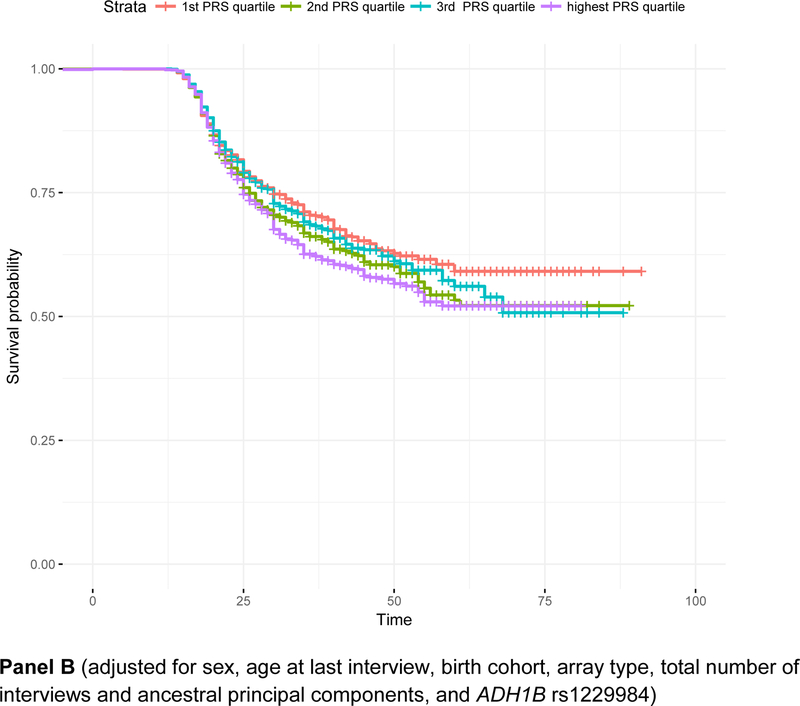
Within the survival analysis framework, alcohol consumption PRS (pT < 0.01) were associated with a 1.11 [95% C.I. 1.06 – 1.16, p = 2.08e-5] hazards of onset of DSM4AD, even after adjustment for covariates. There was no evidence for violation of the proportional hazards assumption. As shown in Figure 1 (Panel A), individuals with scores in the lowest quartile were least likely to have met criteria for DSM4AD, with survival probabilities being higher particularly in those aged 35 and older. Adjustment for rs1229984, which was strongly associated with decreased risk of onset of DSM4AD [Hazards Ratio (HR) = 0.48, 95% C.I. = 0.37 – 0.63, p = 7.85e-08], had little influence on the effect of the overall PRS [HR = 1.09, 95% C.I. = 1.04 −1.14, p = 2.86e-4].
Figure 1. Survival curves representing the relationship between age of onset of alcohol dependence and quartiles of polygenic risk for alcohol consumption.
Panel A: survival curves adjusted for sex, age at last interview, birth cohort, array type, total number of interviews and ancestral principal components; Panel B: adjusted for covariates in Panel A as well as for rs1229984 in ADH1B. Panel A (adjusted for sex, age at last interview, birth cohort, array type, total number of interviews and ancestral principal components) Panel B (adjusted for sex, age at last interview, birth cohort, array type, total number of interviews and ancestral principal components, and ADH1B rs1229984)
As expected, sex was strongly associated with all aspects of problem drinking, including DSM-4AD and DSM5AUDSX (p < 2e-16; higher likelihood of problem drinking in males). Family history was positively associated with all drinking measures except for alcohol use. Age at last interview was associated with DSM-4AD, DSM5AUDSX, and the reduction/cessation comparison of those with no lifetime AUD (Group A) vs. those with lifetime AUD and current symptoms or no symptoms but high-risk drinking (Group B), even after accounting for birth cohort effects. Mean PRS scores did not differ as a function of sex, age, or family history (p > 0.1). Interactions between the PRS and sex, age at final interview and family history were non-significant (p > 0.05), suggesting uniformity of effect sizes across these groups (Table 4).
Table 4. Main effects and PRS interactions with sex, age, and family history of alcohol use disorders for alcohol dependence, AUD symptom count, alcohol use, maximum drinks, and SRE-T.
Beta (SE), and p-value are reported.
| DSM-IV Alcohol Dependence (DSM4AD: PRS pT < 0.01) | DSM-5 AUD Symptom Count (DSM5AUDSX: PRS pT < 0.5) | Alcohol Use (PRS pT < 0.2) | Maximum Drinks (MAXD: PRS pT < 0.1) | Self-rating of effects of ethanol (SRE-T: PRS pT < 0.1) | Reduction/Cessation of problem drinking$ (Reduction: PRS pT < 0.1) | |
|---|---|---|---|---|---|---|
| Sex | −0.986 (0.069), p < 2e-16 | −1.569 (0.082), p < 2e-16 | −1.029 (0.120), p<2e-16 | −0.702 (0.020), p<2e-16 | −0.490 (0.018), p<2e-16 | −0.241 (0.014), p<2e-16 |
| Sex*PRS | −0.015 (0.069), p = 0.833 | 0.149 (0.080), p = 0.064 | 0.099 (0.125), p = 0.428 | 0.032 (0.020), p = 0.103 | 0.021 (0.016), p = 0.185 | 0.008 (0.012), p = 0.485 |
| Age | 0.035 (0.007), p = 3.79e-07 | 0.025 (0.009), p = 0.003 | 0.020 (0.011), p = 0.084 | 0.002 (0.002), p = 0.258 | −0.002 (0.002), p = 0.401 | 0.002 (0.002), p = 0.131 |
| Age*PRS | −0.006 (0.007), p = 0.371 | −0.003 (0.008), p = 0.753 | 0.005 (0.011), p = 0.670 | 7.28e-04 (0.002), p = 0.710 | 8.19e-4 (0.002), p = 0.601 | −0.001 (0.001), p = 0.242 |
| Family history | 0.505 (0.084), p = 1.57e-09 | 0.666 (0.099), p = 1.96e-11 | 0.127 (0.126), p = 0.314 | 0.133 (0.024), p = 4.09e-08 | 0.080 (0.021), p = 1.69e-4 | 0.086 (0.017), p = 2.54e-07 |
| Family history*PRS | −0.040 (0.082), p = 0.630 | 0.102 (0.095), p = 0.280 | 0.123 (0.130), p = 0.344 | 0.009 (0.023), p = 0.708 | 0.017 (0.020), p = 0.385 | −0.004 (0.016), p = 0.812 |
as in Table 3, reported ∆R2 is for the comparison between those with no lifetime AUD vs. those with lifetime AUD and current symptoms or no symptoms but high-risk drinking
DISCUSSION
The GWAS summary statistics used here (Clarke et al., 2017) are derived from simple questionnaire-based items related to typical consumption of various types of alcoholic beverages (e.g., beer, wine, liquor). Such assessments are amenable to collection in very large samples such as the UK Biobank, and meta-analyses of GWAS data for such simple alcohol measures have succeeded in locus discovery (Clarke et al., 2017; Jorgenson et al., 2017; Liu et al., 2019; Schumann et al., 2016). It is unclear whether all loci identified using these indices of typical drinking will overlap with the genetic variants that contribute to other drinking milestones and features, including alcohol dependence. Not only does alcohol dependence involve high levels of alcohol consumption, it includes a significant loss of control over drinking, drinking to ameliorate negative mood states, impairment of interpersonal and vocational functioning, as well as a general transition from impulsive to compulsive use (Koob & Kreek, 2007). One prior GWAS noted that the genetic correlation between alcohol consumption and dependence was modest and variable (Walters et al., 2018). Another recent study that draws on the UK Biobank data suggests that indices of recent problem drinking (e.g., failed to do what was expected because of drinking) were genetically correlated with psychopathology, while indices of more moderate alcohol consumption were not (and in some instances were negatively correlated) (Sanchez-Roige et al., 2018). The present study provides evidence that the aggregated genetic effects related to typical alcohol consumption in an older volunteer cohort do predict significant (albeit very modest) proportions of variance in various indices of drinking, including problem drinking, over and above the effects of rs1229984. However, in the current analyses the strongest associations occurred with a broad measure of alcohol use. This is not entirely surprising, since the alcohol consumption PRS is based on typical weekly intake, which itself is partly derived from frequency of drinking.
Five findings are noteworthy. First, alcohol consumption PRS predicted a very modest proportion of variance (< 0.6%) in DSM IV alcohol dependence (DSM4AD) and DSM 5 AUD symptomatology (DSM5AUDSX). This estimate is relatively consistent with the extent to which PRS tend to explain cross-trait variance. For instance, a recent study examined the extent to which alcohol consumption PRS (derived from a smaller meta-analysis; Schumann et al., 2016) predicted recent drinking (alcohol consumption and AUDIT scores) in a sample of 5,456 participants aged 18–83 years (Mies et al., 2018); at the most predictive thresholds, alcohol consumption PRS predicted 0.11% of the variance in recent drinking. A similar analysis of the Avon Longitudinal Study of Parents and Children (ALSPAC) found that an 89-SNP risk score (derived from literature searches) predicted up to 0.66% of the variance in typical alcohol consumption (Taylor et al., 2016). Despite this consistency, the AUC estimate suggests that the consumption PRS minimally (but significantly) improves classification for DSM4AD. These findings point to the extremely high polygenicity underlying alcohol intake and problem drinking such that even the aggregated effects of SNPs explain only modest proportions of variance. These findings also suggest that there may be only modest genetic overlap between the range of normal alcohol consumption and problematic drinking phenotypes.
Second, despite being a quantitative index of alcohol consumption, maximum number of drinks consumed in a single 24-hour period (MAXD) was less effectively predicted by the alcohol consumption PRS (R2 ≤ 0.31%). MAXD is genetically correlated with problem drinking (Agrawal et al., 2009; Grant et al., 2009) and correlates well with quantitative indices of tolerance (Kendler et al., 2012; Schuckit et al., 2008). The lower prediction might be related to MAXD being potentially influenced by a single episode of heavy drinking and thus, not indicative of either typical or problem drinking. Likewise, total scores on the subjective ratings of ethanol (SRE-T) was not predicted by alcohol consumption PRS. SRE-T is an index of alcohol sensitivity and has primarily been studied as a predisposing factor for and predictor of later problem drinking (Schuckit & Smith, 2013). Individuals with higher SRE-T scores demonstrate lower level of response to alcohol, potentially via dampening of neural and physiological pathways in response to alcohol (Schuckit, 2018; Schuckit et al., 2008). There are currently no published GWAS of SRE-T, although a meta-analysis of a related but etiologically distinct trait representing subjective ratings of ethanol during the first five times that alcohol was consumed (SRE-5) did not find loci previously associated with alcohol consumption or dependence to be related to it (Edwards et al., 2018). Thus, even though SRE is heritable (Kalu et al., 2012) and related to alcohol consumption, its genetic underpinnings may be quite different from those related to alcohol intake.
A third notable observation is that the alcohol consumption PRS explained the most variance for alcohol use (R2 ≤ 1.39%), which represents drinking at least once a month for six consecutive months at some point during the lifetime and is ubiquitously endorsed in this sample (89.9%); this is likely also common in the UK Biobank sample. In fact, the addition of the PRS significantly improved the AUC for alcohol use, compared to a model of covariates only. It is likely that alcohol consumption in the UK Biobank represents normative patterns of drinking and does not sufficiently index problem drinking. Thus, in COGA, PRS derived from such an index of normative alcohol consumption was most closely related to an equally heterogeneous measure of drinking that included normative (and problem) drinkers. We were unable to examine typical alcohol consumption as it was not assessed in COGA in a similar manner as UK Biobank. Nonetheless, our results suggest that in this high-risk sample, PRS from large-scale GWAS of alcohol consumption were more strongly associated with propensity to alcohol use than with measures of problem drinking or disorder.
Fourth, we found little evidence for shared genetic overlap between weekly alcohol consumption and reduction/cessation of problematic drinking. Prior studies have identified candidate variants related to treatment-dependent remission (e.g., (Karpyak et al., 2014)) and emerging evidence suggests high sibling concordance for abstinent remission (McCutcheon et al., 2017), suggesting heritable influences on the transition from active AUD to low-risk drinking and abstinence. However, while PRS distinguished those without a lifetime history of DSM-5 AUD from those with an active diagnosis or high-risk drinking, they were unrelated to low risk drinking or abstinence in those with a lifetime history of DSM-5 AUD. This could be partially due to a lack of power: only 16% of the individuals were in the category of low-risk drinking and abstinence. This finding could also suggest that even though there is polygenic overlap between alcohol consumption and severity of AUD (e.g., DSM5AUDSX), and that individuals who successfully reduce their problem drinking might have a less severe form of the disorder, the genetic propensity that extends beyond severity and specifically predicts cessation is distinct. One might speculate that genetic influences on remission relate less to genetic liability to alcohol intake and more to aspects of socio-economic status (e.g., (Trim et al., 2013)), personality characteristics, as well as other comorbid psychiatric disorders (Lopez‐Quintero et al., 2011). Future studies should further explore the heritability of cessation and its co-heritability with other drinking measures.
Fifth, the addition of rs1229984 in the models attenuated the effect of the PRS, although only modestly for alcohol use. While rs1229984 exerts one of the strongest effect sizes observed for psychiatric and behavioral traits (Edenberg & McClintick, 2018), other loci related to alcohol dependence are expected to have noticeably more modest effects (Walters et al., 2018). In our analyses, the conditional effect of rs1229984 (i.e., R2 for rs1229984 when including PRS in the model) was greater for DSM4AD and DSM5AUDSX (explaining 0.49 – 1.13% of the variance) than the conditional effect of the PRS after adjusting for rs1229984 (0.40 – 0.42% of variance explained). In fact, the addition of PRS to the model had negligible impact on the variance in DSM4AD and DSM5AUDSX already explained by rs1229984, highlighting this variant’s robust effect on problem drinking. In contrast, rs1229984 did not substantially reduce the variance in alcohol use associated with the PRS; rather, the alcohol consumption PRS explained a greater percent of the variance in alcohol use (1.22%) than rs1229984 genotype (0.49%). This suggests that, despite being an aggregate polygenic index of variants with small individual effect sizes, the alcohol consumption PRS are superior predictors of alcohol use compared to rs1229984 alone.
One prior study suggested the polygenic prediction is maximized when the discovery and target cohort have similar demographic characteristics and ascertainment strategies (Savage et al., 2018). Thus, it is also possible that the variance associated with the PRS is greater in the subset of COGA that is demographically matched to the UK Biobank participants (e.g., older age). However, age did not appear to moderate the association between the PRS and alcohol-related outcomes in the current study, nor was there support for absence of proportionality of hazards in the survival model, suggesting homogeneity of effects across age. Relatedly, even though family history was a strong predictor of problem drinking, we found no evidence that it moderated the effect of the PRS in our sample. This might be due to the partially high-risk nature of the COGA sample. Despite differences between the characteristics of the discovery sample and COGA, we detected appreciable variance in alcohol use.
Some limitations of this sample are also noteworthy. First, we restricted our analyses here to individuals of European American descent because the discovery GWAS was limited to individuals of European ancestry, and cross-ancestral predictions have several limitations (Martin et al., 2017). Second, despite being fairly large and predominantly ascertained for families with many alcohol dependent individuals, the COGA sample size may still be underpowered to detect the very modest percentages of variance explained by common SNPs for alcohol-related traits (Bogdan et al., 2018; Clarke et al., 2017; Gratten et al., 2014; Sanchez-Roige et al., 2017; Walters et al., 2018). In particular, although certain types of interaction (e.g. cross-over effects) do not necessitate significant main effects, we were likely under-powered to test such interactions with PRS where a main effect of PRS was undetectable.
While large-scale population-based studies, such as the UK Biobank, are valuable in their potential to identify multiple risk loci for easily-collectible phenotypes, such as alcohol consumption, and contribute to the development of well-powered PRS, they may not adequately capture the genetic factors that contribute to problem drinking, which itself is a multi-faceted phenotype. Until there are similarly large-scale GWAS conducted in samples ascertained for problem drinking, the amount of genetic overlap between alcohol consumption and problematic drinking behaviors remains unclear.
ACKNOWLEDGMENTS
COGA: The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud; Y. Liu); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); Department of Biomedical and Health Informatics, The Children’s Hospital of Philadelphia; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); J. McClintick, L. Wetherill, X. Xuei, D. Lai, S. O’Connor, M. Plawecki, S. Lourens (Indiana University); G. Chan (University of Iowa; University of Connecticut); J. Meyers, D. Chorlian, C. Kamarajan, A. Pandey, J. Zhang (SUNY Downstate); J.-C. Wang, M. Kapoor, S. Bertelsen (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon, S. Saccone (Washington University); J. Salvatore, F. Aliev, B. Cho (Virginia Commonwealth University); and Mark Kos (University of Texas Rio Grande Valley). A. Parsian and H. Chen are the NIAAA Staff Collaborators.
We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
This research is also supported by MH109532 (ECJ, AA, HJE) and F32AA027435 (ECJ).
Footnotes
COMPETING INTERESTS
J.I.N. is an investigator for Janssen and Assurex.
References
- 1000 Genomes Project Consortium. (2015). A global reference for human genetic variation. Nature, 526(7571), 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerbo E, Sullivan PF, Vilhjalmsson BJ, Pedersen CB, Mors O, Børglum AD, Hougaard DM, Hollegaard MV, Meier S, Mattheisen M, Ripke S, Wray NR, Mortensen PB (2015). Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry, 72(7), 635–641. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, Madden PA, Todorov A, Trull T, Bucholz KK, Todd RD, Sher K, Heath AC (2009). Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. Journal of Studies on Alcohol and Drugs, 70(2), 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT, Heath AC, Chassin L (2011). Developing a genetically informative measure of alcohol consumption using past-12-month indices. Journal of Studies on Alcohol and Drugs, 72(3), 444–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. (2000). American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision. American Psychiatric Publishing, Inc. [Google Scholar]
- American Psychiatric Association. (2013). American Psychiatric Association: Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition. American Psychiatric Publishing, Inc. [Google Scholar]
- Bartoń K (2012). MuMIn: multi-model inference: R package See: http://cran.r-project.org/web/packages/MuMIn/index.html.
- Begleiter H, Reich T, Hesselbrock V, Poriesz B, Li TK, Schuckit MA, Edenberg HJ, Rice JP (1995). The collaborative study on the genetics of alcoholism. Alcohol Health & Research World, 19(3), 228–237. [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI, Porjesz B, Saccone NL, Schuckit MA, Tischfield J, Wang J, Foroud TM, Rice JP, Edenberg HJ (2012). ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Molecular Psychiatry, 17(4), 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan R, Baranger DAA, Agrawal A (2018). Polygenic Risk Scores in Clinical Psychology: Bridging Genomic Risk to Individual Differences. Annual Review of Clinical Psychology, 14(1), 119–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr, Reich T, Schmidt I, Schuckit MA (1994). A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. Journal of Studies on Alcohol, 55(2), 149–158. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, McCutcheon VV, Agrawal A, Dick DM, Hesselbrock VM, Kramer JR, Kramer JR, Kuperman S, Nurnberger JI Jr, Salvatore JE, Schuckit MA, Bierut LJ, Foroud TM, Chan G, Hesselbrock M, Meyers JL, Edenberg HJ, Porjesz B (2017). Comparison of Parent, Peer, Psychiatric, and Cannabis Use Influences Across Stages of Offspring Alcohol Involvement: Evidence from the COGA Prospective Study. Alcoholism: Clinical and Experimental Research, 41(2), 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A Day FR, Loh P-R, ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, Duncan L., Perry JRB, Patterson N, Robinson EB, Daly MJ Price AL Neale BM (2015). An atlas of genetic correlations across human diseases and traits. Nature genetics, 47(11), p.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ, McIntosh AM (2017). Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N= 112 117). Molecular psychiatry, 22(10), 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh P-R, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C (2016). Next-generation genotype imputation service and methods. Nature genetics, 48(10), 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J (2012). Improved whole-chromosome phasing for disease and population genetic studies. Nature methods, 10(1), 5. [DOI] [PubMed] [Google Scholar]
- Dick DM, Meyers JL, Rose RJ, Kaprio J, Kendler KS (2011). Measures of current alcohol consumption and problems: two independent twin studies suggest a complex genetic architecture. Alcoholism: Clinical and Experimental Research, 35(12), 2152–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F (2013). Power and predictive accuracy of polygenic risk scores. PLoS genetics, 9(3), e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ (2007). The Genetics of Alcohol Metabolism: Role of Alcohol Dehydrogenase and Aldehyde Dehydrogenase Variants. Alcohol Research & Health, 30(1), 5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, McClintick JN (2018). Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcoholism: Clinical and Experimental Research, 42(12), 2281–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Deak JD, Gizer IR, Lai D, Chatzinakos C, Wilhelmsen KP, Lindsay J, Heron J, Hickman M, Webb BT, Bacanu S-A, Foroud TM, Kendler KS, Dick DM, Schuckit MA (2018). Meta‐Analysis of Genetic Influences on Initial Alcohol Sensitivity. Alcoholism: Clinical and Experimental Research, 42(12), 2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JD, Agrawal A, Bucholz KK, Madden PAF, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, Whitfield JB, Martin NG, Heath AC (2009). Alcohol consumption indices of genetic risk for alcohol dependence. Biological psychiatry, 66(8), 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Wray NR, Keller MC, Visscher PM (2014). Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nature Neuroscience, 17, 782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Martin NG (1994). Genetic Influences on Alcohol Consumption Patterns and Problem Drinking: Results from the Australian NH&MRC Twin Panel Follow‐up Survey. Annals of the New York Academy of Sciences, 708(1), 72–85. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V (1999). A validity study of the SSAGA‐a comparison with the SCAN. Addiction, 94(9), 1361–1370. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460(7256), 748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C, Choquet H (2017). Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Molecular psychiatry, 22(9), 1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalu N, Ramchandani VA, Marshall V, Scott D, Ferguson C, Cain G, Taylor R (2012). Heritability of Level of Response and Association with Recent Drinking History in Nonalcohol‐Dependent Drinkers. Alcoholism: Clinical and Experimental Research, 36(6), 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpyak VM, Biernacka JM, Geske JR, Jenkins GD, Cunningham JM, Rüegg J, Kononenko O, Leontovich AA, Abulseoud OA, Hall-Flavin DK, Loukianova LL, Schneekloth TD, Skime MK, Frank J, Nöthen MM, Rietschel M, Kiefer F, Mann KF, Weinshilboum RM, Frye MA, Choi D-S (2014). Genetic markers associated with abstinence length in alcohol-dependent subjects treated with acamprosate. Translational psychiatry, 4(10), e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A, Kosinski M, Biecek P (2017). survminer: Drawing Survival Curves using’ggplot2’. R Package Version 0.3, 1. [Google Scholar]
- Keller MC (2014). Gene× environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biological psychiatry, 75(1), 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Prescott CA, Crabbe J, Neale MC (2012). Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Molecular Psychiatry, 17(12), 1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Dick D, Prescott CA (2010). The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcoholism: Clinical and Experimental Research, 34(6), 1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob G, Kreek MJ (2007). Stress, dysregulation of drug reward pathways, and the transition to drug dependence. American Journal of Psychiatry, 164(8), 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler H, Zhou H, Kember R, Smith RV, Justice A, Damrauer S, Tsao PS, Klarin D, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, Gelernter J (2019). Genome-wide Association Study of Alcohol Consumption and Use Disorder in Multiple Populations (N= 274,424). bioRxiv, 527929.
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hinds DA, Hromatka BS, Huber KE, Kleinman A, Litterman NK, McIntyre MH, Mountain JL, Northover CAM, Sathirapongsasuti JF, Sazonova OV, Shelton JF, Shringarpure S, Tian C, Tung JY, Vacic V, Wilson CH, Pitts SJ, Mitchell A, Skogholt AH, Winsvold BS, Sivertsen B, Stordal E, Morken G, Kallestad H, Heuch I, Zwart J-A, Fjukstad KK, Pedersen LM, Gabrielsen ME, Johnsen MB, Skrove M, Indredavik MS, Drange OK, Bjerkeset O, Børte S, Stensland SØ, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga J-J, Huang H, Jang S-K, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJ, Vrieze S (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature genetics, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Quintero C, Hasin DS, De Los Cobos JP, Pines A, Wang S, Grant BF, Blanco C (2011). Probability and predictors of remission from life‐time nicotine, alcohol, cannabis or cocaine dependence: Results from the national epidemiologic survey on alcohol and related conditions. Addiction, 106(3), 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczak SE, Glatt SJ, Wall TJ (2006). Meta-analyses of ALDH2 and ADH1B with alcohol dependence in asians. Psychological Bulletin, 132, 607–621. [DOI] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE (2017). Human demographic history impacts genetic risk prediction across diverse populations. The American Journal of Human Genetics, 100(4), 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon VV, Schuckit MA, Kramer JR, Chan G, Edenberg HJ, Smith TL, Bender AK, Hesselbrock V, Hesselbrock M, Bucholz KK (2017). Familial association of abstinent remission from alcohol use disorder in first-degree relatives of alcohol-dependent treatment-seeking probands. Addiction, 112(11), 1909–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mies GW, Verweij KJH, Treur JL, Ligthart L, Fedko IO, Hottenga JJ, Willemsen G, Bartels M, Boomsma DI, Vink JM (2018). Polygenic risk for alcohol consumption and its association with alcohol-related phenotypes: Do stress and life satisfaction moderate these relationships? Drug and Alcohol Dependence, 183, 7–12. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G (2017). Collider scope: when selection bias can substantially influence observed associations. International Journal of Epidemiology, 47(1), 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Schielzeth H (2012). A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods in Ecology and Evolution, 4(2), 133–142. [Google Scholar]
- National Institute on Alcohol Abuse and Alcoholism. (2004). NIAAA council approves definition of binge drinking. NIAAA Newsletter, 3(3). [Google Scholar]
- Nyholt DR (2004). A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. The American Journal of Human Genetics, 74(4), 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, & Weeks DE (1998). PedCheck: A Program for Identification of Genotype Incompatibilities in Linkage Analysis. The American Journal of Human Genetics, 63(1), 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS (1999). Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. American Journal of Psychiatry, 156(1), 34–40. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007). PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics [DOI] [PMC free article] [PubMed]
- R Core Team. (2017). R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, & Patra J (2009). Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. The Lancet, 373(9682), 2223–2233. [DOI] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, Gray JC, de Wit H, Davis LK, MacKillop J, Palmer AA (2017). Genome‐wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addiction biology, 24(1), 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, the 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, Adams MJ, Howard DM, Edenberg HJ, Davies G, Crist RC, Deary IJ, I AM, and Clarke T-K (2018). Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. American Journal of Psychiatry, 176(2), 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage JE, Salvatore JE, Aliev F, Edwards AC, Hickman M, Kendler KS, Macleod J, Latvala A, Loukola A, Kaprio J, Rose RJ, Chan G, Hesselbrock V, Webb BT, Adkins A, Bigdeli TB, Riley BP, Dick DM (2018), Polygenic Risk Score Prediction of Alcohol Dependence Symptoms Across Population‐Based and Clinically Ascertained Samples. Alcoholism: Clinical and Experimental Research, 42: 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA (2018). A Critical Review of Methods and Results in the Search for Genetic Contributors to Alcohol Sensitivity. Alcoholism: Clinical and Experimental Research, 42(5), 822–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL (2013). Stability of scores and correlations with drinking behaviors over 15 years for the self-report of the effects of alcohol questionnaire. Drug and Alcohol Dependence, 128(3), 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Danko G, Kramer J, Bucholz KK, McCutcheon V, Chan G, Kuperman S, Hesselbrock V, Dick DM, Hesselbrock M, Porjesz B, Edenberg HJ, Nurnberger JI, Gregg M, Schoen L, Kawamura M, Mendoza LA (2018). A 22-Year Follow-Up (Range 16 to 23) of Original Subjects with Baseline Alcohol Use Disorders from the Collaborative Study on Genetics of Alcoholism. Alcoholism: Clinical and Experimental Research, 0(0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Tipp JE (1997). The self‐rating of the effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction, 92(8), 979–988. [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim RS, Heron J, Horwood J, Davis J, Hibbeln J, ALSPAC Study Team. (2008). The self-rating of the effects of alcohol questionnaire as a predictor of alcohol-related outcomes in 12-year-old subjects. Alcohol & Alcoholism, 43(6), 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivières S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proença C, Chambers JC, Clarke T-K, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernández-Medarde A, Foroud T, Freimer NB, Girault J-A, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen A-L, Heath AC, Hesselbrock V, Hofman A, Hottenga J-J, Isohanni MK, Kaprio J, Khaw K-T, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Núñez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tönjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin M-R, Elliott P (2011). Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proceedings of the National Academy of Sciences, 108(17), 7119–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Lepe MS, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke T-K, Enroth S, Fischer K, Hällfors J, Harris SE, Hieber S, Hofer E, Hottenga J-J, Johansson Å, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J, Lyytikäinen L-P, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud A-C, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, Geus E. de, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen A-L, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin M-R, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimäki T, Liu Y, Madden PAF, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, Ridker P, Rose R, Rotter JI, Samani NJ, Schmidt H, Spector TD, Stott D, Strachan D, Tzoulaki I, Harst P. van der, Duijn C.M. van, Marques-Vidal P, Vollenweider P, Wareham NJ, Whitfield JB, Wilson J, Wolffenbuttel B, Bakalkin G, Evangelou E, Liu Y, Rice KM, Desrivières S, Kliewer SA, Mangelsdorf DJ, Müller CP, Levy D, Elliott P (2016). KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA 113(50), 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell T, Zhao J, Panwar S, Roemer A, Naimi T, Chikritzhs T (2016). Do “moderate” drinkers have reduced mortality risk? A systematic review and meta-analysis of alcohol consumption and all-cause mortality. Journal of studies on alcohol and drugs, 77(2), 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow DI, Kuchenbaecker KB, Shah S, Sofat R, Holmes MV, White J, Mindell JS, Kivimaki M, Brunner EJ, Whittaker JC, Casas JP (2016). Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. International journal of epidemiology, 45(5), 1600–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Simpkin AJ, Haycock PC, Dudbridge F, Zuccolo L (2016). Exploration of a polygenic risk score for alcohol consumption: a longitudinal analysis from the ALSPAC cohort. PloS One, 11(11), e0167360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, Grambsch PM (2013). Modeling survival data: extending the Cox model Springer Science & Business Media. [Google Scholar]
- Therneau TM, Lumley T (2015). Package ‘survival.’ R Top Doc, 128. [Google Scholar]
- Trim RS, Schuckit MA, Smith TL (2013). Predictors of Initial and Sustained Remission from Alcohol Use Disorders: Findings from the 30‐Year Follow‐Up of the San Diego Prospective Study. Alcoholism: Clinical and Experimental Research, 37(8), 1424–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, Kendler KS (2015). The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychological Medicine, 45(5), 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen L-S, Clarke T-K, Chou Y-L, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, Foo JC, Fox L, Frank J, Giegling I, Gordon S, Hack LM, Hartmann AM, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Jan Hottenga J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Lai D, Ligthart L, Loukola A, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Meyers JL, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang J-C, Webb BT, Wedow R, Wetherill L, Wills AG, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hinds DA, Huber KE, Kleinman A, Litterman NK, McCreight JC, McIntyre MH, Mountain JL, Noblin ES, Northover CAM, Pitts SJ, Sathirapongsasuti JF, Sazonova OV, Shelton JF, Shringarpure S, Tian C, Tung JY, Vacic V, Wilson CH, Boardman JD, Chen D, Choi D-S, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye MA, Gäbel W, Hayward C, Ising M, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Männistö S, Müller-Myhsok B, Murray AD, Nurnberger JI, Palotie A, Preuss U, Räikkönen K, Reynolds MD, Ridinger M, Scherbaum N, Schuckit MA, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma DI, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AM, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer CJ, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak VM, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes HH, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nöthen MM, Palmer AA, Pedersen NL, Penninx BWJH, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen P-H, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A, (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience, 21(12), 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RA, Koob GF (2014). The development and maintenance of drug addiction. Neuropsychopharmacology, 39(2), 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AM, Kaptoge S, Butterworth AS, Willeit P, Warnakula S, Bolton T, Paige E, Paul DS, Sweeting M, Burgess S, Bell S, Astle W, Stevens D, Koulman A, Selmer RM, Verschuren WMM, Sato S, Njølstad I, Woodward M, Salomaa V, Nordestgaard BG, B Yeap B.B., Fletcher A, Melander O, Kuller LH, Balkau B, Marmot M, Koenig W, Casiglia E, Cooper C, Arndt V, Franco OH, Wennberg P, Gallacher J, de la Cámara AG, Völzke H, Dahm CC, Dale CE, Bergmann MM, Crespo CJ, van der Schouw YT, Kaaks R, Simons LA, Lagiou P, Schoufour JD, Boer JMA, Key TJ, Rodriguez B, Moreno-Iribas C, Davidson KW, Taylor JO, Sacerdote C, Wallace RB, Quiros JR, Tumino R, Blazer DG, Linneberg A, Daimon M, Panico S, Howard B, Skeie G, Strandberg T, Weiderpass E, Nietert PJ, Psaty BM, Kromhout D, Salamanca-Fernandez E, Kiechl S, Krumholz HM, Grioni S, Palli D, Huerta JM, Price J, Sundström J, Arriola L, Arima H, Travis RC, Panagiotakos DB, Karakatsani A, Trichopoulou A, Kühn T, Grobbee DE, Barrett-Connor E, van Schoor N, Boeing H, Overvad K, Kauhanen J, Wareham N, Langenberg C, Forouhi N, Wennberg M, Després J-P Cushman M, Cooper JA, Rodriguez CJ, Sakurai M, Shaw JE, Knuiman M, Voortman T, Meisinger C, Tjønneland A, Brenner H, Palmieri L, Dallongeville J, Brunner EJ, Assmann G, Trevisan M, Gillum RF, Ford I, Sattar N, Lazo M, Thompson SG, Ferrari P, Leon DA, Smith GD, Peto R, Jackson R, Banks E, Di Angelantonio E, Danesh J (2018). Risk thresholds for alcohol consumption: combined analysis of individual-participant data for 599 912 current drinkers in 83 prospective studies. The Lancet, 391(10129), 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2014). Global status report on alcohol and health, 2014 World Health Organization. [Google Scholar]
- Wray NR, Lee SH, Mehta D, Vinkhuyzen AA, Dudbridge F, Middeldorp CM (2014). Research review: polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry, 55(10), 1068–1087. [DOI] [PubMed] [Google Scholar]
- Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM (2013). Pitfalls of predicting complex traits from SNPs. Nature Reviews Genetics, 14, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youden WJ (1950). Index for rating diagnostic tests. Cancer, 3(1), 32–35. [DOI] [PubMed] [Google Scholar]