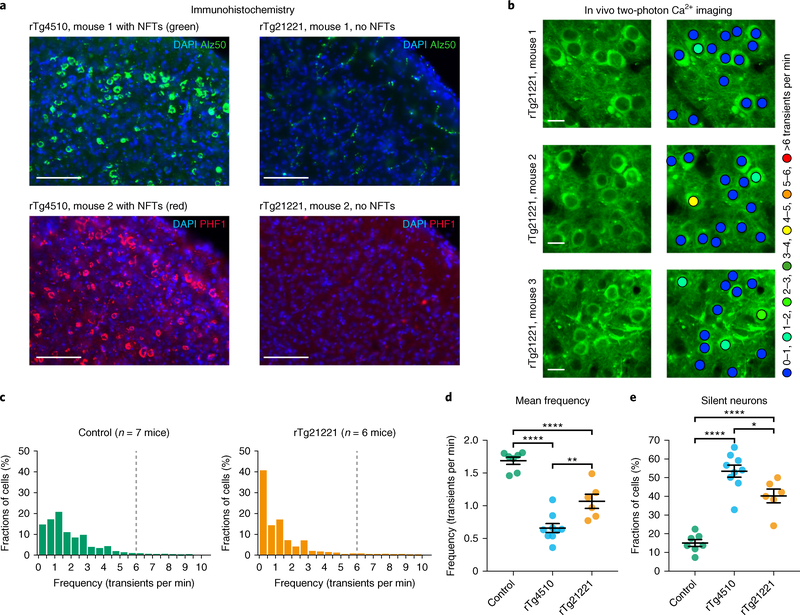
Fig. 2 |. NFTs are not required for neuronal silencing.
a, Coronal sections showing many Alz50-positive (top, green) and PHF1-positive (bottom, red) NFTs in the cortex of rTg4510 mice (n = 4 mice, 8–10 sections per mouse were analyzed), but not in rTg21221 mice (n = 4 mice, 5–12 sections per mouse were analyzed). Nuclei are visualized with DAPI (blue). Scale bars, 100 μm. b, In vivo two-photon fluorescence images of GCaMP6f-expressing (green) layer 2/3 neurons and corresponding activity maps from three example rTg21221 mice illustrating the marked silencing of many neurons. Scale bars, 10 μm. c, Frequency distributions of all recorded neurons in wild-type controls (left panel, green, n = 1,705 neurons in 7 mice) and rTg21221 mice (right panel, orange, n = 1,021 neurons in 6 mice). d, Mean frequency of silent neurons. Controls: 1.69 ± 0.05 transients per min; rTg4510: 0.66 ± 0.07 transients per min; rTg21221: 1.07 ± 0.11 transients per min. F(2,19) = 48.43, P = 3.47 × 10−8. All post hoc multiple comparisons between genotypes were significant: P = 2.01 × 10−8 for controls vs. rTg4510, P = 1.00 × 10−4 for controls vs. rTg21221, P = 0.0038 for rTg4510 vs. rTg21221. e, Fractions of silent neurons. Controls: 15.05 ± 1.87%; rTg4510: 53.48 ± 3.24%; rTg21221: 40.25 ± 3.64%. F(2,19) = 42.94, P = 8.94 × 10−8. All post hoc multiple comparisons between genotypes were significant: P = 5.55 × 10−8 for controls vs. rTg4510, P = 7.85 × 10−5 for controls vs. rTg21221, P = 0.0179 for rTg4510 vs. rTg21221. The data for the controls and rTg4510 mice are the same as in Fig. 1. Each solid circle represents an individual animal and all error bars reflect the mean ± s.e.m; the differences between genotypes were assessed by one-way ANOVA followed by Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, ****P < 0.0001.