Figure 1. DEAD-box proteins unwind duplexes non-processively via local strand separation.
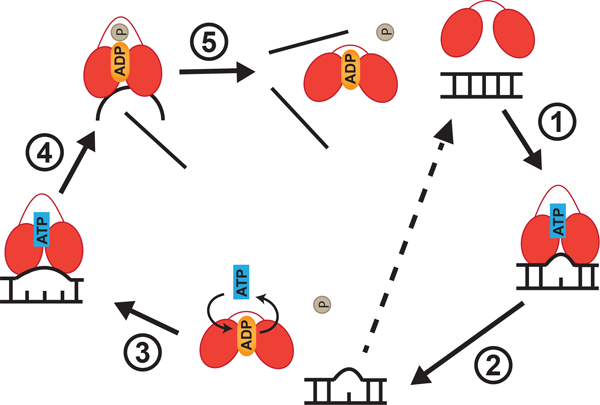
Schematic representation of the unwinding cycle of a prototypical DEAD-box RNA helicase. Lines represent RNA strands and the two ovals represent the two RecA-like domains that are connected by a flexible linker. In the absence of any nucleotide and RNA, the two RecA-like domains are farther apart and exhibit a flexible “opened, unproductive” conformation. During unwinding, the two RecA-like domains come closer together to form a “closed, productive” conformation upon binding to the double-stranded RNA (dsRNA) and ATP (step 1). Closing of the two domains bends one strand of the dsRNA and results in local duplex destabilization (~ 6 bp). Duplexes longer than 6 bp require multiple cycles of unwinding. ATP hydrolysis and inorganic phosphate release convert the two RecA-like domains back to the “opened” conformation (step 2). This causes dissociation of the helicase core from the partially opened dsRNA. The partially opened dsRNA can potentially snap back to produce a non-productive unwinding cycle (dotted arrow) or is subjected to another round of local duplex destabilization (step 3). Another round of local duplex destabilization happens after the ADP is exchanged to ATP in the DEAD-box protein or a new ATP-bound DEAD-box protein recognizes the partially opened dsRNA. This allows the DEAD-box protein to fully disrupt the partially opened duplex. Upon ATP hydrolysis, the ADP-Pi bound DEAD-box protein still associates with the bent strand whereas the non-bent strand is released (step 4). The bent strand is eventually dissociated from the DEAD-box protein once the inorganic phosphate is released (step 5) (Figure adapted from Rudolph and Klostermeier 2015).
