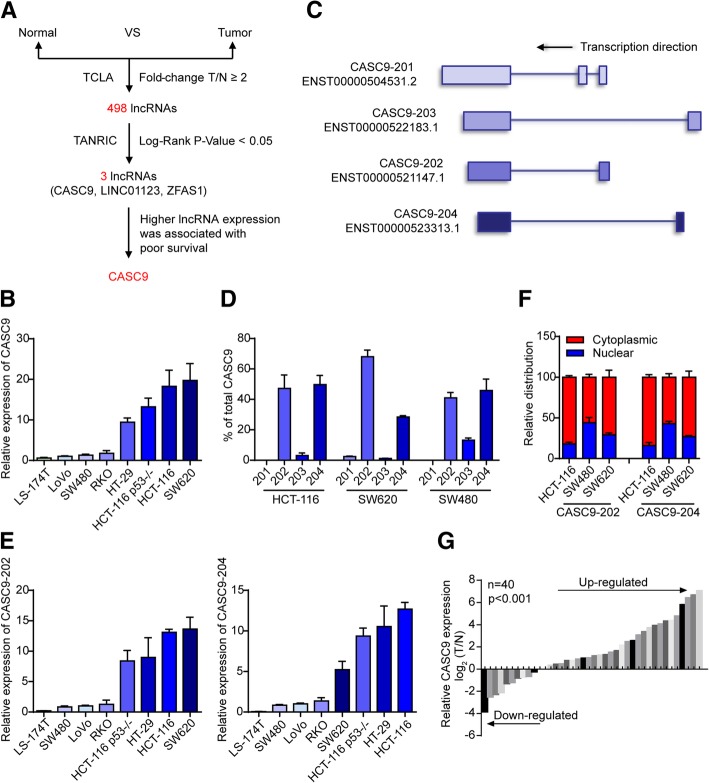
Fig. 1.
Identification of CASC9 as an lncRNA upregulated in CRC. a Schematic representation of the process used to identify upregulated lncRNAs with significant prognostic values in COAD. b Expression of CASC9 in CRC cell lines was quantified by RT-qPCR. β-Actin mRNA was used as an internal control. c Exon maps for four transcript variants of CASC9 with Ensembl IDs. d RNA expression analysis of the four transcript variants of CASC9 in HCT-116, SW620, and SW480 cells by RT-qPCR. e RNA expression analysis of the two most abundant CASC9 transcript variants, CASC9–202 (left) and CASC9–204 (right), in CRC cell lines. f Subcellular localization of CASC9–202/204 was analyzed by RT-qPCR upon biochemical fractionation of HCT-116, SW480, and SW620 cells. GAPDH mRNA was used as a control for cytoplasmic transcripts, NEAT1 was used as a control for nuclear transcripts. g RT-qPCR analysis of relative CASC9 expression in 40 primary CRC tissues and paired normal tissues. The expression of CASC9 was normalized to that of β-actin. The data are presented as the mean ± s.d.