Extended Data Fig. 5. Design and biochemical characterization of GIFNs.
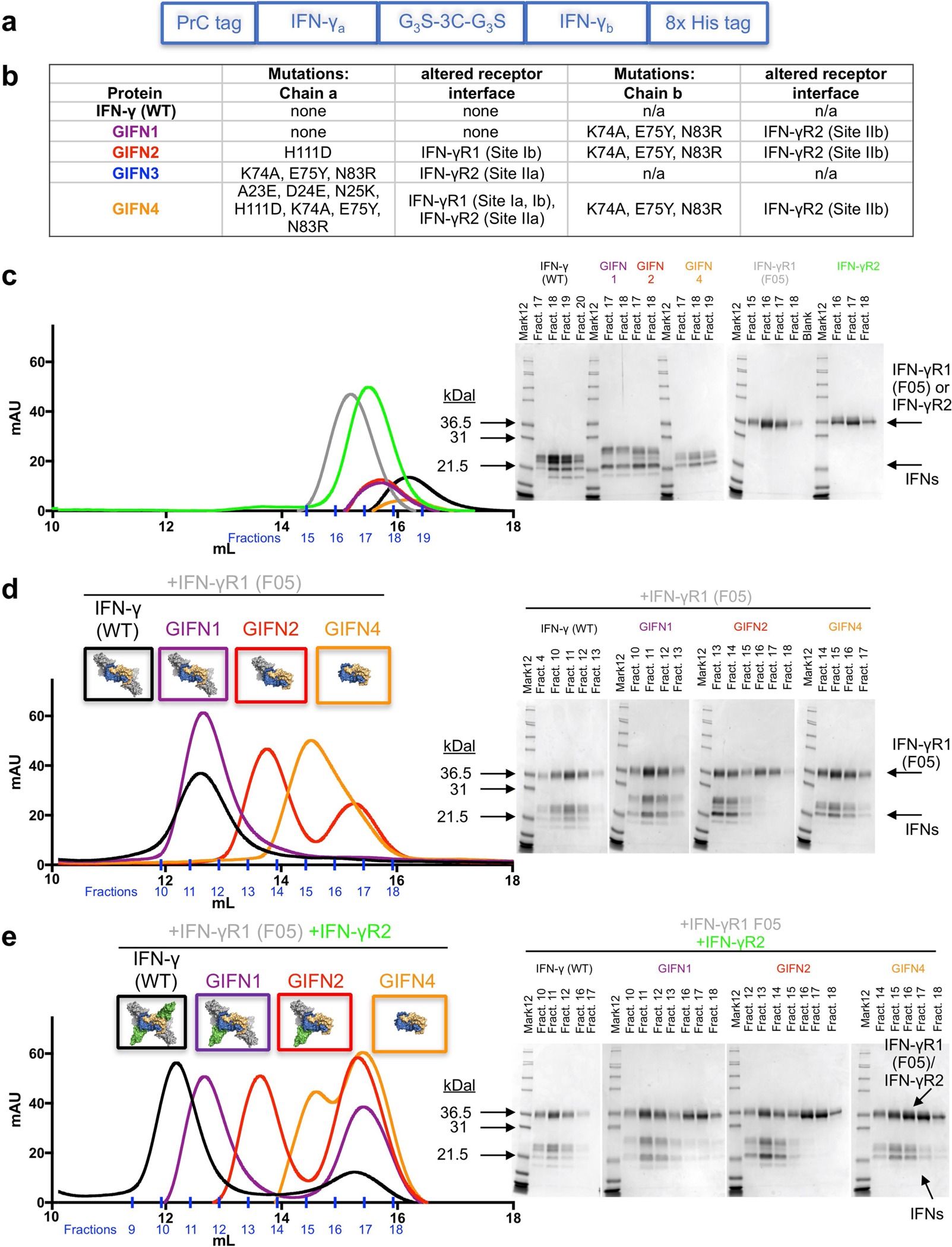
a, Diagram showing the strategy for expression and purification of heterodimeric IFNγ variants. The asymmetric variants containing three tags were expressed as single polypeptides in Hi5 insect cells. The proteins were first harvested from the secreted medium with a C-terminal 8× His tag. Proteins eluted from nickel resin were then treated with 1:100 (by mass) human rhinovirus 3C protease for 24 h at 4 °C to cleave the 3C protease tag. The 3C tag is flanked by Gly-Gly-Gly-Ser motifs at both ends (G3S-3C-G3S) to ensure accessibility of the protease site between the two chains of IFNγ. The cleaved proteins were then purified using the N-terminal protein C-tag and a final 8× His tag purification to ensure isolation of the correctly paired heterodimeric IFNγ proteins. b, Table of mutations for each of the GIFN proteins, indicating the affected receptor binding sites. GIFN2, one IFNγR1 and one IFNγR2 binding in cis; GIFN3, two IFNγR1 molecules bound; GIFN4, reduced affinity for both IFNγR1 and IFNγR2. c, SEC profiles and SDS–PAGE gel fractions for 200 μg wild-type IFNγ (black) or equal molar quantities of GIFN1 (purple), GIFN2 (red), GIFN4 (orange), IFNγR1 F05 (grey), and IFNγR2 (green). Individual proteins have been purified and analysed by SEC at least three times. d, e, To determine the receptor-binding properties of the GIFN proteins, the shifts in the SEC profiles and gels were compared relative to the wild-type protein as described for c except IFNs were mixed with equimolar quantities of IFNγR1 F05 (d) or equimolar quantities of both IFNγR1 F05 and IFNγR2 (e). These data are from single experiments except the wild-type experiments which were performed at least three times.
