Extended Data Fig. 3. Quantification of IFNγR2(T168N) glycoforms by mass spectroscopy.
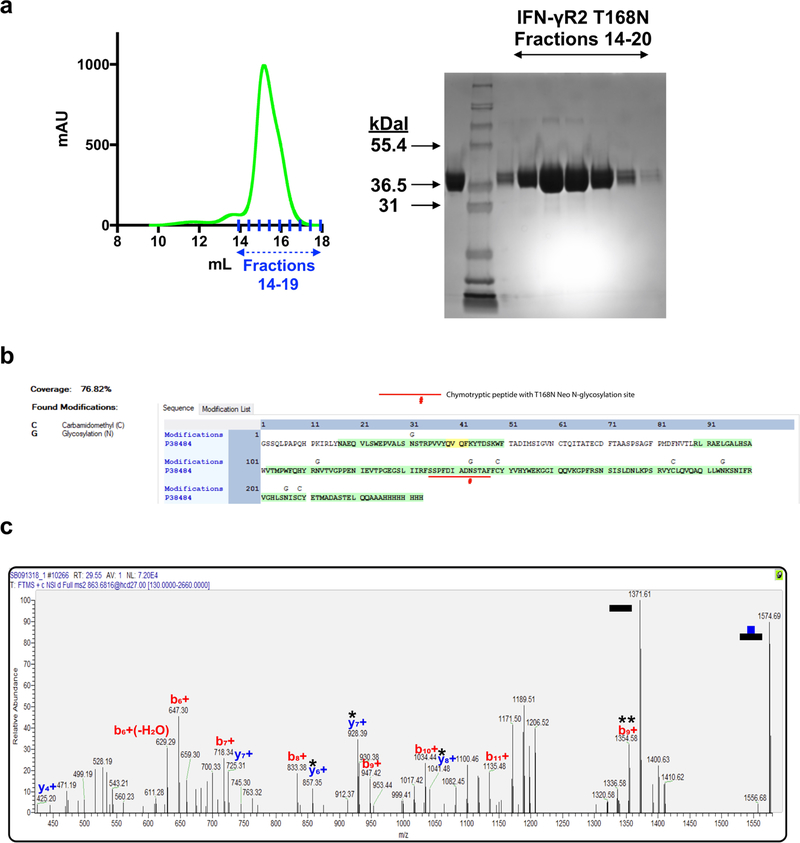
a, The mutant IFNγR2(T168N) protein was expressed in HEK293S GnTI− cells and purified by SEC. The SEC profile is shown (left) with the corresponding fractions on SDS–PAGE (right). Lane 1 shows the sample loaded on the SEC column, lane 2 shows the Mark 12 protein ladder, and lanes 3–9 are fractions 14–20. Data are representative of at least 3 biologically independent experiments. b, The protein coverage map shows sequence coverage of 76.82% for the entire IFNγR2(T168N) protein including the peptide of interest, containing N168, which is underlined. This peptide was detected as a glycopeptide with several glycoforms as quantified in Fig. 3d. Mappings highlighted in green indicate high confidence with a false discovery rate (FDR) below 1% and yellow indicates a FDR of 1–5%. Carbamidomethyl (C) and glycosylation (G) sites are indicated above the site of modification. Confidence levels were determined as previously described38. c, MS2 spectra confirming that the ion used for the EICs shown in Fig. 3d is the peptide SSPFDIADNSTAF from IFNγR2(T168N) modified with a HexNAc2Hex5 glycan attached to N168. The data shown are for a single experiment.
