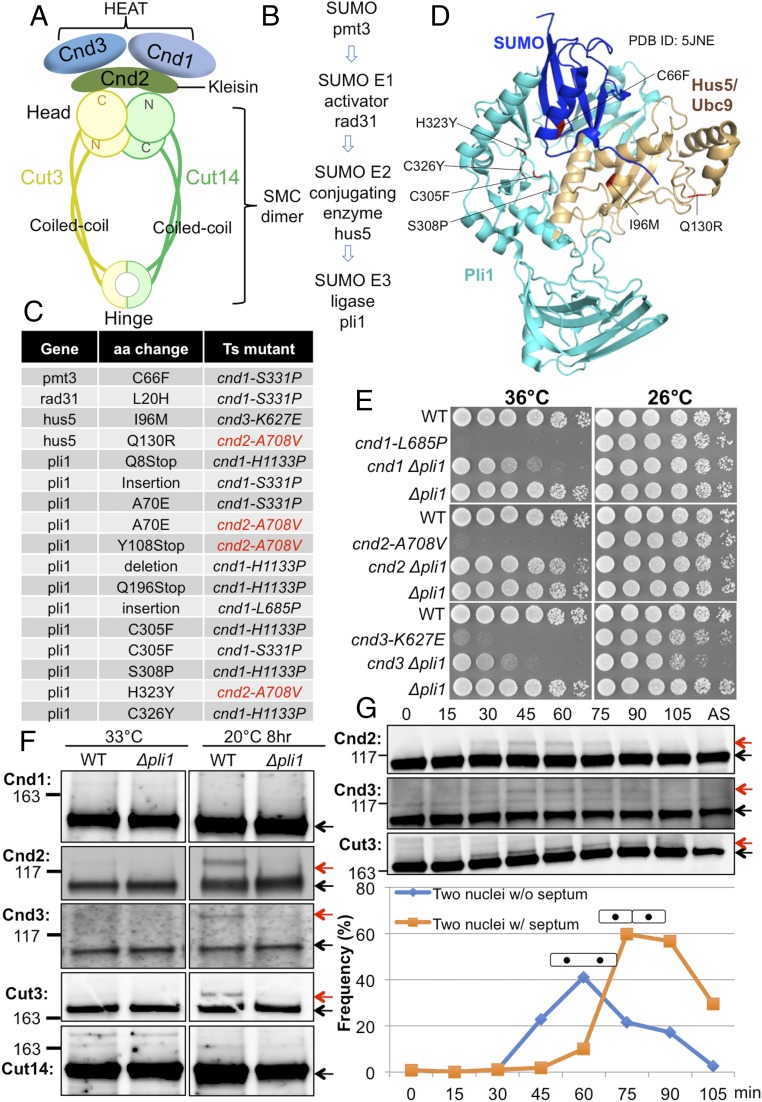
Fig. 1.
Loss of SUMOylation suppresses ts cnd1, cnd2, and cnd3 mutations. (A) Arrangement of the subunits in the fission yeast condensin complex. Cut3/SMC4 and Cut14/SMC2 are SMC proteins, while Cnd1, Cnd2/kleisin, and Cnd3 are the three non-SMC proteins that bind to the SMC head domain. (B) Genes identified as suppressors for cnd mutants form the SUMOylation pathway. (C) SUMOylation mutations that suppressed cnd1, cnd2, and cnd3 ts mutants. The majority of them are in the SUMO E3 ligase gene pli1. (D) Location of suppressor mutations in the 3D structure of SUMO, Hus5/Ubc9, and Pli1 (PDB ID code 5JNE) (Materials and Methods). (E) Spot tests showed extragenic suppression of cnd1, cnd2, and cnd3 mutants by deletion of the pli1 gene, which encodes SUMO E3 ligase. WT, wild type. (F) Immunoblotting of WT or ∆pli1 in the background of the β-tubulin mutant nda3-KM311 cultured at 33 °C (asynchronous culture) and 20 °C (restrictive temperature, 8 h; cells were arrested at prometaphase) was performed. The upper SUMO bands (red arrows) were detected for Cnd2, Cnd3, and Cut3 in the WT and were abolished in the deletion mutant ∆pli1. (G) Block and release experiment was done using the ts cdc25-22 mutant. These cells were blocked in late G2 phase and released synchronously into mitosis by a temperature shift from the restrictive temperature, 36 °C, to the permissive temperature, 26 °C. Aliquots were taken every 15 min for immunoblotting and measurement of the septation index. Cnd2, Cnd3, and Cut3 clearly produced upper bands (red arrows) only during mitosis. Cells of two nuclei without (w/o) septum are mitotic cells, while cells with (w/) septum are postmitotic, but before cytokinesis. The protein bands, that are not SUMOylated, were indicated by black arrows in F and G.