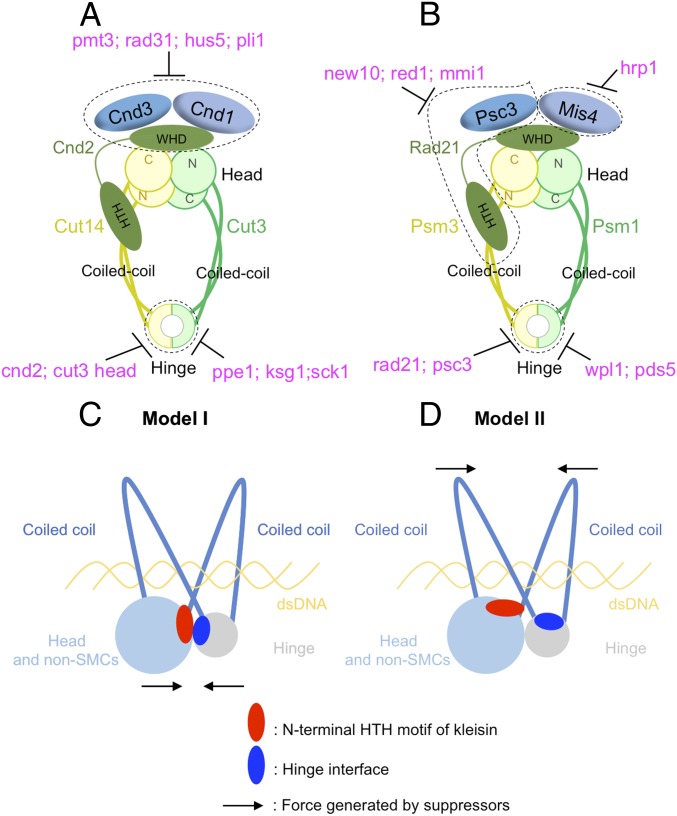
Fig. 6.
Condensin and cohesin are regulated differently, but they may adopt a similar organization in which the hinge and head interact. (A) Summary of condensin’s suppression by SUMOylation pathway mutants (pmt3, rad31, hus5, and pli1), kinase or phosphatase mutants (ppe1, ksg1, and sck1), and condensin mutants (cnd2 and cut3 head mutations). (B) Summary of cohesin’s suppression by RNA elimination pathway mutants (new10, red1, and mmi1), chromatin-remodeling factor mutants (hrp1), cohesin-releasing factor mutants (wpl1 and pds5), and cohesin non-SMC mutants (rad21 and psc3) (text). Two models were proposed in C and D to explain SMC hinge interface mutants’ suppression by mutations in the N-terminal HTH motif of kleisins. (C) SMC hinge interfaces and the N-terminal HTH motif of kleisins may interact directly to form arched coiled coils, which hold and release chromosomal DNA. SMC hinge interface mutations may impair head–hinge interaction, and suppressors in the N-terminal HTH motif of kleisins rescue the interaction. (D) SMC hinge interfaces and the N-terminal HTH motif of kleisins may not directly interact, but they both regulate coiled-coil orientation. SMC hinge interface mutations may widen the coiled-coil angle, thereby impairing the capacity of the coiled coils to hold chromosomal DNA. Suppressors in the N-terminal HTH motif of kleisins rescue the DNA-binding ability of the coiled coils.