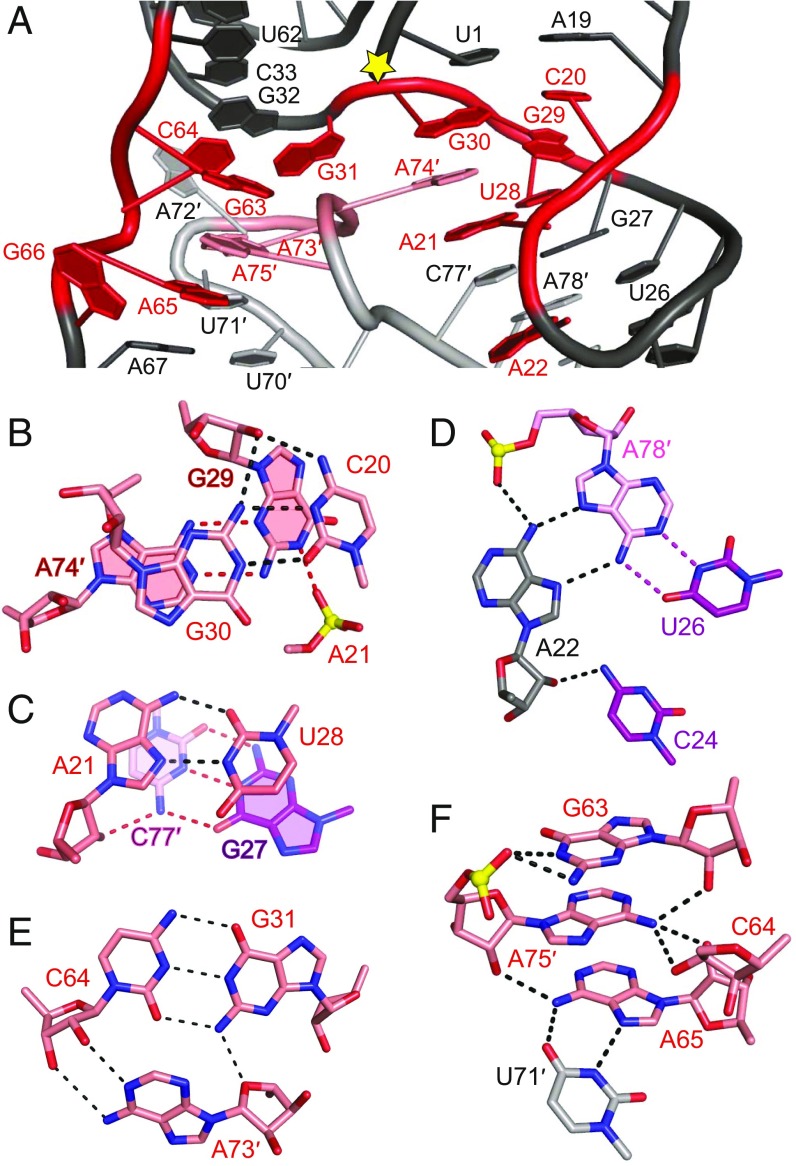
Fig. 2.
Structural alignment of highly conserved residues in the hatchet ribozyme. (A) Highly conserved residues (in red) are brought into proximity near the cleavage site (labeled with a yellow star) through pairing and hydrogen bonding interactions in the tertiary structure. (B) C20 forms a trans-Watson–Crick base pair with G30 adjacent to the terminal part of stem P1, in which G30 adopts a C2′-endo sugar pucker. The sugar edge of G29 formed a sheared pairing interaction with the Watson–Crick edge of A74′, with the Watson–Crick edge of G29 forming additional hydrogen bonds with the nonbridging phosphate oxygen of A21, resulting in a stable interaction plane. The 2′-OH of G29 is pointed outwards from the plane and forms hydrogen bonds with the above G30-C20 base pair. Notably, both the sugar pucker of G29 and A74 adopted C2′-endo conformations. (C) The Hoogsteen edge of A21 forms a trans-pairing interaction with the Watson–Crick edge of U28; the 2′-OH of A21 forms one hydrogen bond with the adjacent stacked base pair G27-C77′ from stem P2. The sugar pucker of A21 adopts a C2′-endo conformation. (D) Highly conserved A22 forms a major groove-aligned base triple interaction with the Watson–Crick U26-A78′ base pair. The 2′-OH of A22 forms an additional hydrogen bond with 4-NH2 of C24. The sugar pucker of A22 adopts a C2′-endo conformation. (E) The minor groove-aligned base triple A73′•(G31-C64) involves highly conserved residues A73′, G31, and C64. (F) In molecule A′ of the HT-GAAA structure, the base G63 hydrogen bonds with the phosphate oxygen of A75′. The 6-NH2 of A75′ forms hydrogen bonds with the sugars of three residues G63, C64, and A65. The 2′-OH of A75′ hydrogen bonds with 6-NH2 of A65. A65 formed a cis-Hoogsteen Watson–Crick base pair with U71′. Note that the dashed lines indicate distances <3.5 Å and their number can exceed the possible number of hydrogen bonds formed by an atom.