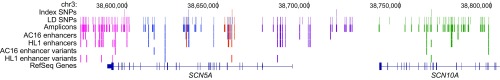
Fig. 1.
Genomic map of common variants at the QTi-associated SCN5A-SCN10A GWAS locus on chromosome 3p22.2. A 224-kb genomic segment is annotated with tracks, showing (from top) the five independent GWAS hits (Index SNPs); all common (MAF > 5%) SNPs in 1000 Genomes Project European ancestry samples in moderate to high LD (r2 > 0.3) with the five index SNPs (LD SNPs); amplicons encompassing the LD SNPs that were cloned and evaluated in reporter assays (Amplicons); amplicons that were cis-regulatory enhancers by in vitro reporter assays in the human cardiomyocyte cell line AC16 (AC16 enhancers) and the mouse cardiomyocyte cell line HL1 (HL1 enhancers); the cis-regulatory enhancers that displayed significant allelic difference in reporter activities in AC16 (AC16 enhancer variants) and HL1 (HL1 enhancer variants); and the protein-coding SCN5A and SCN10A (RefSeq) genes. The five independent GWAS hits are marked in color (pink, blue, brown, purple, and green) in the Index SNPs track, while features in other tracks are color-coded based on the highest LD with these five index variants. The genomic map was generated using custom tracks in the UCSC Genome Browser.