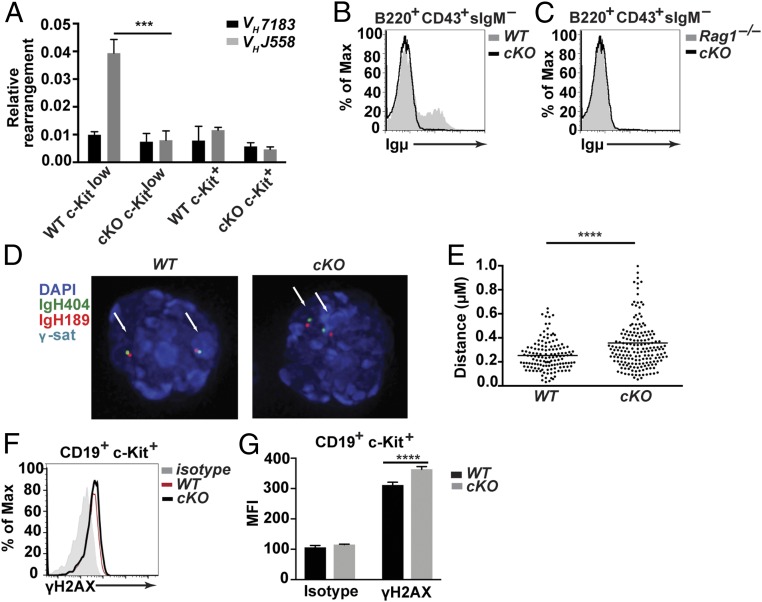
Fig. 4.
Loss of CHD4 results in reduced VH to DJH rearrangements and increased DNA damage in pro-B cells. (A) qPCR analysis of rearranged proximal (VH7183) and distal (VHJ558) gene segments from genomic DNA isolated from sorted c-Kit+ pro-B cells (Lin–CD19+c-Kit+CD25–IgM–IgD–) and c-Kitlow pro-B cells (Lin–CD19+c-KitlowCD25–IgM–IgD–) from bone marrow of 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ mice (n = 3) and WT littermate controls (n = 3). (B) Flow cytometry of cytoplasmic Ig µ-expression in sorted B220+CD43+sIgM– B cells isolated from bone marrow from 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ (n = 2) and WT (n = 2) mice. (C) Flow cytometry of cytoplasmic Ig µ-expression in sorted B220+CD43+sIgM– B cells isolated from bone marrow from 4- to 6-wk-old Chd4fl/flCd79a-CreTg/+ (n = 2) and a Rag1 knockout mouse (n = 1). Data are derived from one experiment. (D) The 3D-FISH confocal images of hybridized probes in CD19+c-Kit+ pro-B cell (as described above) nuclei isolated from bone marrow of Chd4fl/flCd79a-CreTg/+ and WT mice. The IgH404 proximal probe (RP23-404D8) was labeled with Alexa Fluor 488 (AF488); the IgH189 distal probe (RP23-189H12) was labeled with AF568; major satellite repeats (γ-sat) were conjugated to AF647; and nuclei were counterstained with DAPI. (E) Distributions of spatial distances (in micrometers) separating BAC probes, located at each end of the Igh locus (IgH404 vs. IgH189) in pro-B cells isolated from bone marrow of Chd4fl/flCd79a-CreTg/+ (n = 92 cells) and WT (n = 69 cells). Each dot represents an individual allele. (F) Representative flow cytometry and (G) MFI of c-Kit+ pro-B cells stained for intracellular γ-H2AX from bone marrow of Chd4fl/flCd79a-CreTg/+ (n = 4) and WT (n = 3) mice. The asterisks indicate statistical significance compared with the WT littermate controls as unpaired, two-tailed Student’s t test with equal SD, Mann–Whitney rank test or two-way ANOVA with Sidak’s multiple comparisons test. **P < 0.0005, ****P < 0.0001. All data are representative of at least three independent experiments.