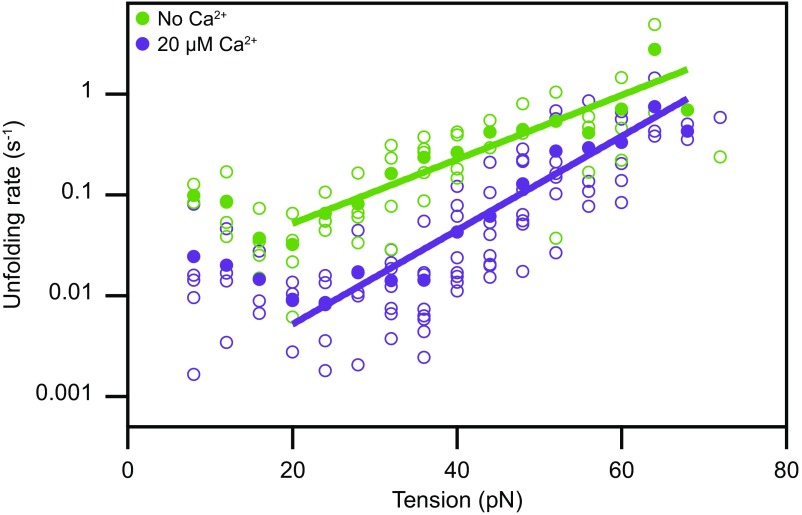
Fig. 4.
Tension-dependent unfolding rates of a single cadherin domain. Assuming that all domains of PCDH15 are similar and unfold independently, we estimate the rate at which individual EC domains unfold as a function of tension. Domains unfold much more readily in the absence of Ca2+ (green) than at a physiological Ca2+ concentration of 20 µM (purple). The filled circles represent the means for eight molecules at a Ca2+ concentration of 20 µM and for five molecules in the absence of Ca2+. The outlined circles, which represent the data for individual molecules, provide an estimate of the data’s spread. The solid lines are fits of Bells’ model (48) to the data. For 20 µM Ca2+, the unfolding rate with no force is k0 = 0.0006 ± 0.0002 s−1, and the transition-state distance is x‡ = 0.44 ± 0.04 nm; in the absence of Ca2+, these values are k0 = 0.012 ± 0.006 s−1 and x‡ = 0.30 ± 0.04 nm.