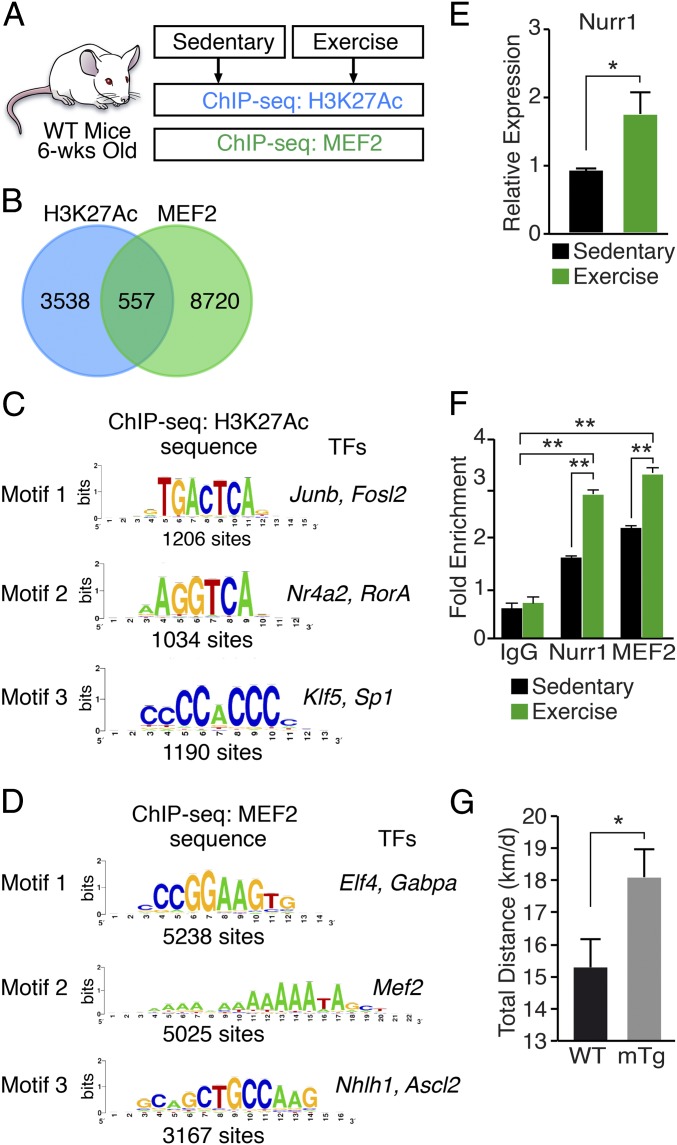
Fig. 1.
Chromatin analysis in response to exercise. (A) Schematic of chromatin immunoprecipitation experiments using tibialis anterior muscle from sedentary mice or mice following 8 wk of voluntary wheel running beginning at 6 wk of age. (B) Venn diagram showing overlap of 557 exercise-responsive genes associated with ChIP-seq peaks for H3K27Ac and MEF2 in skeletal muscle. For H3K27Ac ChIP-seq, 3,955 enhancer peaks were associated with 4,095 genes. For MEF2 ChIP-seq, 10,032 peaks were associated with 9,277 genes. (C) De novo motif discovery from peaks of the H3K27Ac ChIP-seq reads. The three most abundant sequence motifs are shown. (D) De novo motif discovery from peaks of the MEF2 ChIP-seq reads. The three most abundant sequence motifs are shown. (E) Nurr1 mRNA normalized to 18S RNA in quadriceps muscle of sedentary mice, and mice following 8 wk of voluntary wheel running. n = 6. (F) ChIP-seq assays showing binding of NURR1 and MEF2 to the Glut4 promoter in quadriceps muscle of sedentary mice, and mice following 8 wk of voluntary wheel running. Antibodies against NURR1 or MEF2 were used in ChIP-seq assays for the Glut4 promoter. Graphs display mean quantification of ChIP-seq data (percentage of input) normalized to IgG control. IgG control, n = 3. Sedentary mice, n = 6. Exercised mice, n = 6. (G) WT and Nurr1-mTg mice were subjected to a voluntary wheel-running regimen for 8 wk. Nurr1-mTg mice showed enhanced physical performance, measured by total distance run in km/d. n = 6. Data are represented as mean ± SEM. *P < 0.05, **P < 0.005.