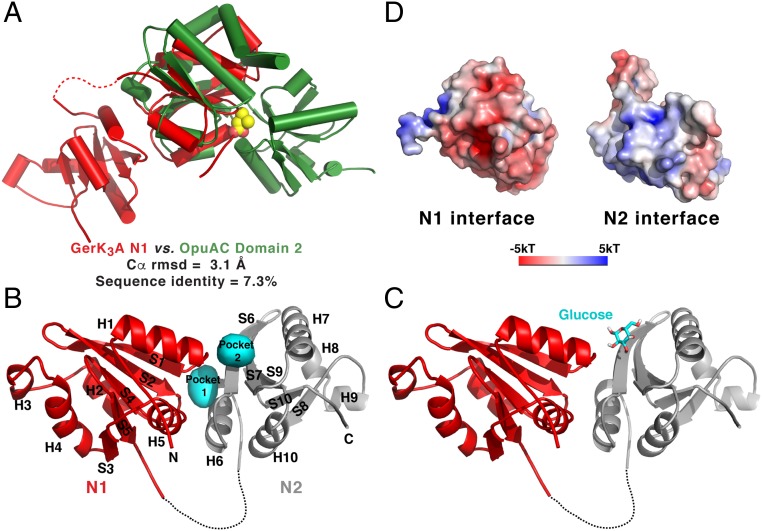
Fig. 3.
The structure of GerK3ANTD is similar to those of PeBP proteins. (A) Superimposition of the N1 domain of GerK3ANTD (red) and the domain 2 of OpuAC (green; PDB ID code 2B4L). The bound ligand for OpuAC, glycine betaine, is shown in space-filling representation. (B) Molecular surface representation of the two interfacial pockets (cyan) between the N1 (red) and N2 (gray) domains of GerK3ANTD defined by HOLLOW (66). (C) One of the top-ranked poses of the glucose molecule determined by AutoDock Vina is shown as licorice sticks in the presumed binding site in GerK3ANTD. (D) Molecular surface representation of the interface between the N1 and N2 domains of GerK3ANTD colored according to the local electrostatic potential (−5 kT to +5 kT), calculated with the program ABPS (65).