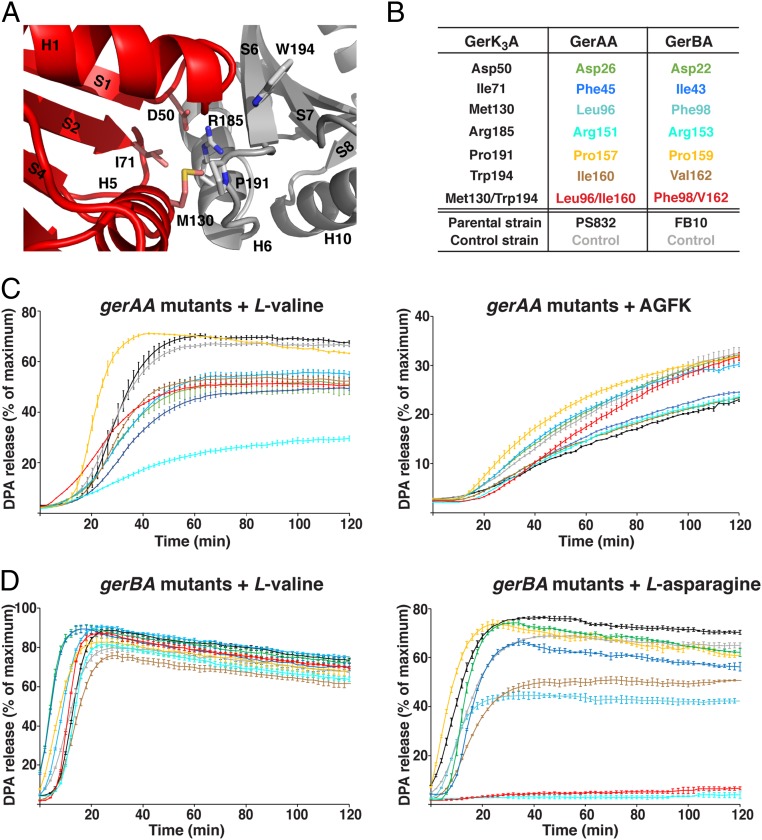
Fig. 4.
Effects of B. subtilis gerAA and gerBA putative interface mutations on spore germination. (A) Close-up view of the N1–N2 interface in GerK3ANTD with selective interfacial residues of the N1 (pink) and N2 (gray) domains shown as licorice sticks. Numbers of secondary structure elements are labeled as in Fig. 1B. (B) The list of the putative N1–N2 interfacial WT residues in B. subtilis GerAA and GerBA proteins that were subjected to alanine substitution, and their equivalent residues in GerK3ANTD. Note that the WT B. subtilis gerAA and gerBA genes were cloned and transformed into PS832 or FB10, respectively, and the resulting strains were considered and labeled as control strains in this experiment to assess any effects of the cloning alone. (C) DPA release from spores with the predicted gerAA interface mutations in the PS832 background in the presence of 10 mM l-valine (Left) or 10 mM AGFK (Right). (D) DPA release from spores with the predicted gerBA interface mutations in the FB10 background in the presence of 10 mM l-valine (Left) or 10 mM l-asparagine (Right). The colors of the germination curves correspond to the mutations listed in B. For each strain, the percentage of DPA release was normalized against the RFU readings obtained from the same spores boiled in water. Data represent means ± SD for at least three independent measurements.