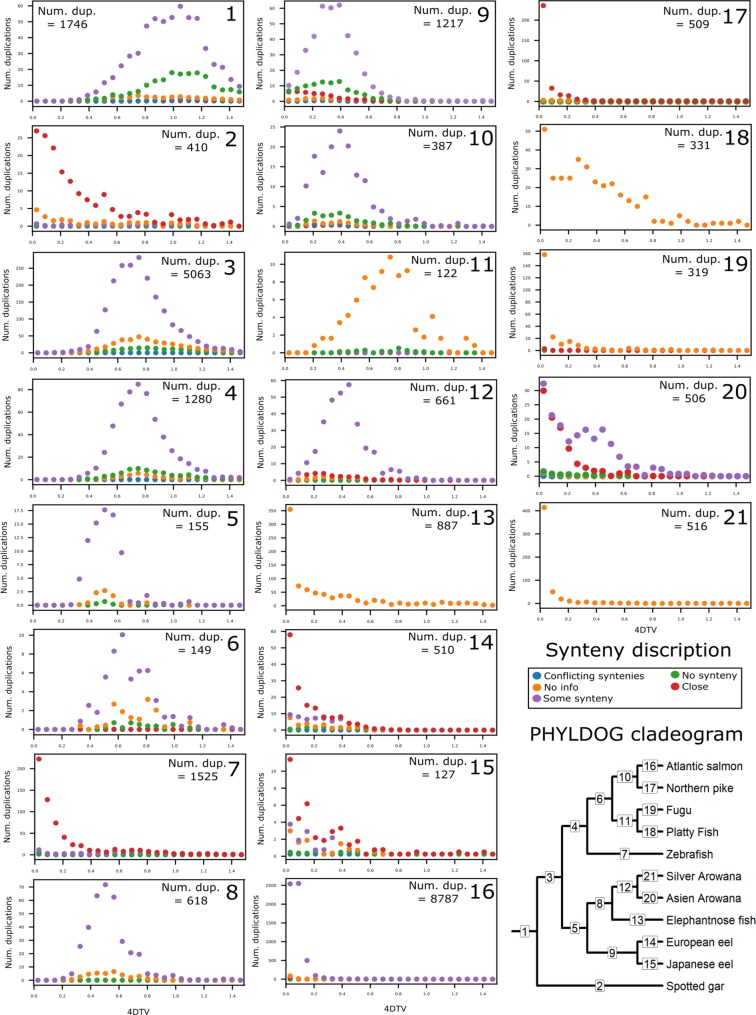
Fig 4. 4dTv and synteny distributions of duplications per branch of the PHYLDOG species tree.
Quantity, 4dTv and synteny distributions of duplications assigned to each branch of the PHYLDOG species tree. Each panel represents the branch with the corresponding number in the cladogram in the bottom right-hand corner. Species included in this study are: European eel (Anguilla anguilla), Japanese eel (Anguilla japonica), zebrafish (Danio rerio), northern pike (Esox lucius), spotted gar (Lepisosteus oculatus), fugu (Takifugu rubripes), platyfish (Xiphophorus maculatus), Atlantic salmon (Salmo salar), elephantnose fish (Gnathonemus petersii), Asian arowana (Scleropages formosus) and silver arowana (Osteoglossum bicirrhosum). The synteny types are the following: close (), duplicated genes are close in the genome; some synteny (), paralogues of genes found close to one duplicate are also found close to the other duplicate; no synteny (), less than two paralogeus for other genes are found close to both paralogue duplicates; no information (), the duplicated genes are located in small scaffolds with too few genes close by; conflicting syntenies (), different synteny classifications found in the genomes of the different species affected by the duplication.