Fig. 3. Comparative assessment of genome assemblies and chromosomal evolution across the Lepidoptera.
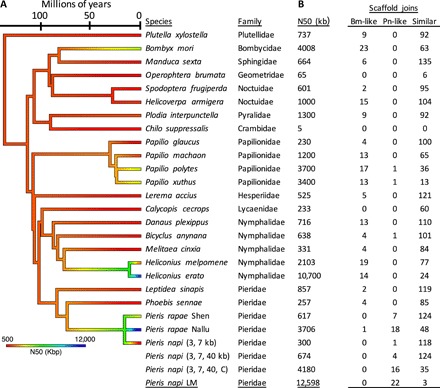
(A) A time-calibrated phylogeny of the currently available lepidopteran genomes (n = 24), with branches colored by the N50 of their assemblies and time in million years ago. (B) Table of the genome assembly N50 for each species, followed by estimates of their chromosomal similarity relative to B. mori versus P. napi. In each scaffold of each genome, SCOs were identified that were shared with B. mori and P. napi. Then, we quantified the number of times a scaffold contained SCOs derived from >1 chromosome of B. mori, but from a single P. napi chromosome (Pnapi-like scaffold), or vice versa (Bmori-like scaffold), or both from one chromosome (similar) (fig. S3 and note S9). Three additional P. napi genomes are included, representing the original Allpaths assembly that used only 3- and 7-kb MP libraries [Pieris napi (3, 7 kb)], the Allpaths assembly after an additional round of scaffolding with 40-kb MP libraries [Pieris napi (3, 7, and 40 kb)], and then the 40-kb scaffolded assembly scaffolded a third time and error-corrected with HiC libraries and the HiRise pipeline [Pieris napi (3, 7, and 40 kb)]. Higher Pn-like values indicate support for a P. napi–like chromosomal structure (see note S11 for more details). For a complementary BLAST-like approach, see fig. S9.
