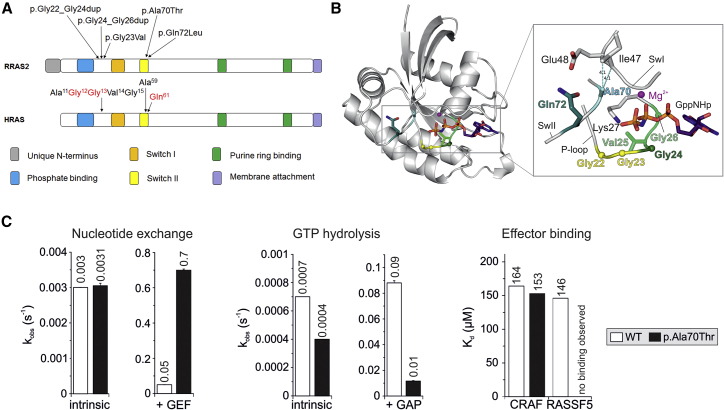
Figure 2.
RRAS2 Structure and Location and Functional Impact of Noonan Syndrome-Causing Variants
(A) Schematic representation of RRAS2 and HRAS proteins. Conserved motifs critical for tight guanine nucleotide binding and hydrolysis, and position of the disease-causing RRAS2 variants are illustrated together with the homologous residues of HRAS. The three residues representing the mutational hotspots of oncogenic HRAS mutations are shown in red.
(B) Structural modeling of RRAS2 variants. A structural model of the active GTP-bound RRAS2 protein highlights the relative position of the disease-causing missense or insertion mutations. All RRAS2 mutations affect residues that are located in the nucleotide binding active site region, which contains integral elements involved in GDP/GTP binding, GTP hydrolysis, and interactions with regulators (GEFs and GAPs) and effectors.
(C) Biochemical assessment of RRAS2p.Ala70Thr. RRAS2WT and RRAS2p.Ala70Thr proteins were biochemically characterized regarding their nucleotide exchange (left), GTP hydrolysis (middle), and effector binding (right) properties. The nucleotide exchange reaction was measured in the absence (intrinsic) and in the presence of the catalytic RASGEF domain of mouse RASGRF1, while the catalytic activity of the GTPase was assessed in the absence (intrinsic) and in the presence of the p120 RASGAP GAP domain. The RAS-binding and RAS association domains of CRAF and RASSF5 were used to evaluate the binding behavior of the RRAS2p.Ala70Thr mutant to RAS effectors. Overall, the data indicate that the p.Ala70Thr change leads to an accumulation of the protein in its GTP-bound active state, resulting to an increased signaling activity. The missense change, however, is predicted to differentially impact on the diverse downstream signaling pathways.