Figure 3.
Biochemical and Functional Characterization of Noonan Syndrome-Causing RRAS2 Variants
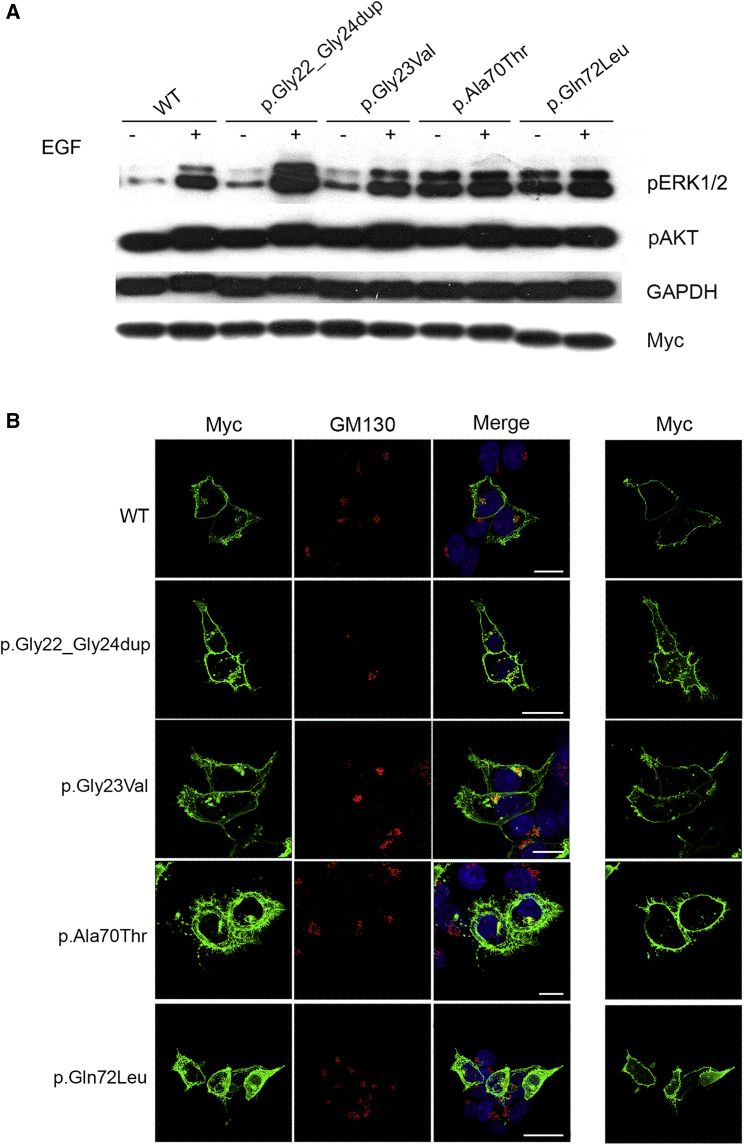
(A) ERK and AKT phosphorylation assays. HEK293T cells were transfected with the indicated Myc-tagged RRAS2 constructs. Following starvation (18 h) and EGF stimulation (30 ng/mL for 15 min), ERK and AKT phosphorylation levels were evaluated using a mouse monoclonal anti-phospho-p44/42 ERK (Thr202/Tyr204) antibody and a rabbit polyclonal anti-phospho-AKT (Ser473) antibody, respectively. To assess myc-RRAS2 protein levels, 20 μg of total lysates were immunoblotted with a mouse monoclonal anti-Myc antibody. Membranes were re-probed with mouse monoclonal anti-GAPDH antibody for protein normalization. Representative blot of three performed experiments are shown.
(B) RRAS2 subcellular localization showed by confocal laser scanning microscopy (CLSM) observations (left). Assays were performed on HEK293T cells starved overnight and stained with an anti-Myc mouse monoclonal antibody, followed by goat anti-mouse Alexa Fluor-488 (green), and an anti-GM130 (Golgi marker) rabbit polyclonal antibody, followed by goat anti-rabbit Alexa Fluor-594 (red). Nuclei are visualized by DAPI staining (blue). Co-localization areas were detected in yellow. CLSM observation were also performed at the basal level of cells to show the distinctive pattern of adhesion-like structures and cytoskeletal rearrangement in cells expressing the RRAS2 mutants (right). In all panels, bars correspond to 21 μm.