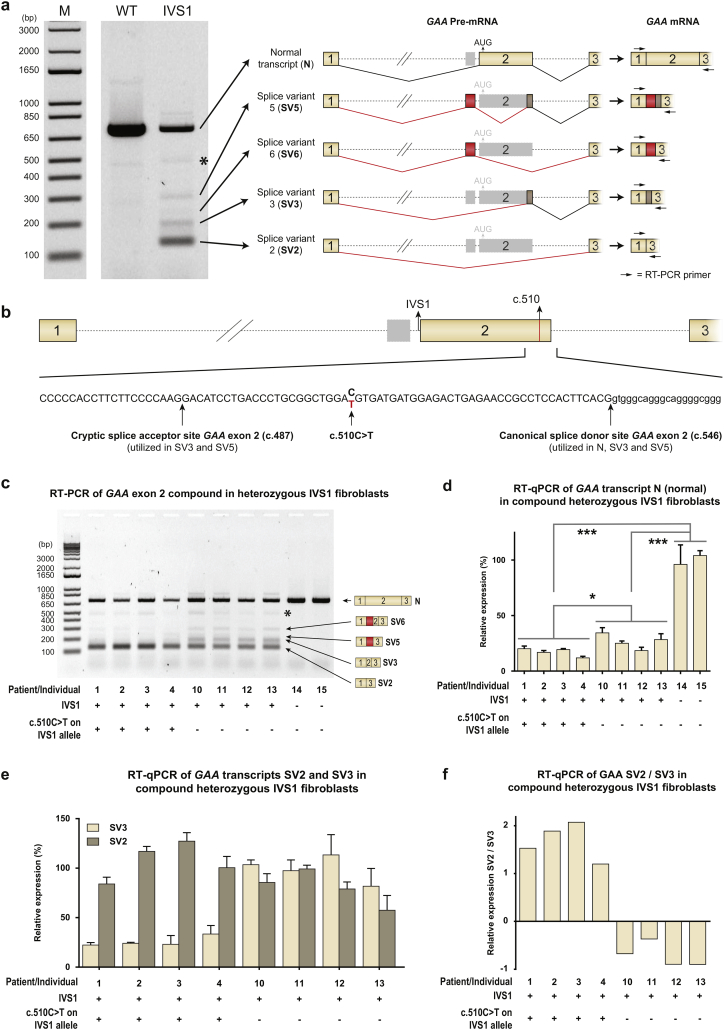
Fig. 3.
c.510C>T worsens splicing outcome in compound heterozygous IVS1 patients. (A) Left: RT-PCR analysis of GAA exons 1–3 of fibroblasts from a healthy control (WT) and a compound heterozygous IVS1 patient (IVS1). The second allele in this patient did not produce mRNA. M indicates the DNA size marker in base pairs (bp). Right: Cartoons of the major splice products. Numbered boxes represent canonical exons. Unnumbered boxes represent facultative exons or parts of exons: a pseudoexon in intron 1 (in red) or the C-terminal part of exon 2, which is derived from utilization of the cryptic splice site at c.487 (in brown). Dotted lines represent introns. Continuous lines represent splicing events. *: structural variant (see Suppl. Fig. S4). (B) Position of c.510C>T in GAA exon 2. Positions of the cryptic and canonical splice sites of exon 2 are also indicated. Numbered boxes represent canonical exons. The unnumbered gray box represent the pseudoexon in intron 1. Dotted lines represent introns. (C) RT-PCR analysis of GAA exon 1 to 3 in fibroblasts from compound heterozygous IVS1 patients with and without c.510C>T. Splice products N, SV6, SV5, SV3 and SV2 are indicated on the right. *: structural variant (see Suppl. Fig. S4). The patients or individuals analyzed are indicated and listed in Supplementary Tables S2 and S3. (D) As (C), but now analyzed using RT-qPCR for the wildtype splice product N. (E) As (C), but now analyzed using RT-qPCR for the aberrant splice products SV2 and SV3. (F) Ratio of SV2/SV3. In healthy control cells, expression of SV2 and SV3 was too low to allow quantification. Data in d-e represent means ± SD (n= 3 biological replicates). * = p < 0.05, *** = p < 0.001.