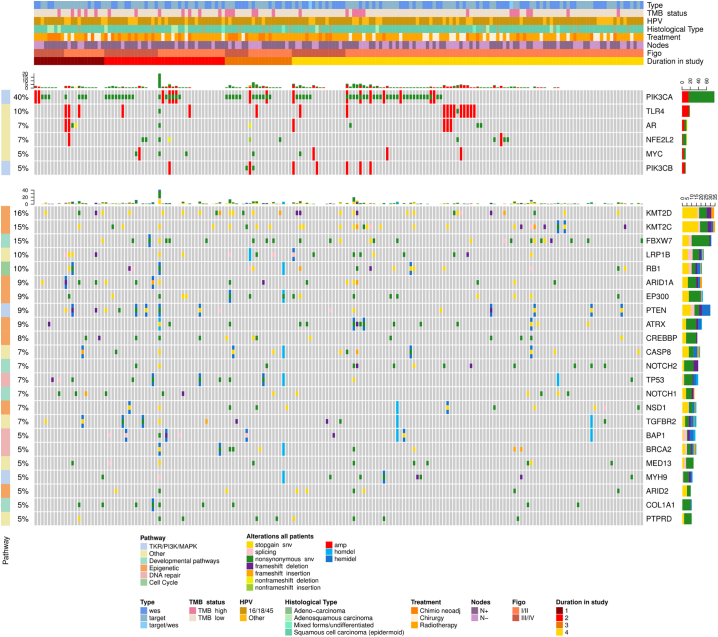
Fig. 1.
Oncoprints are presented, showing the most frequently altered (higher than 5% of the cohort) oncogenes and tumour suppressor genes, combining both small and large-scale variations together with clinical information (WES and target panel combined), using the R package Complex Heatmap (v1.17.1). Each column represents a patient. Key oncogene and tumour suppressor gene alterations are listed by name (right column), by frequency in the population (left column), by alteration type (colour code) by each clinical data detail for each individual patient (heading). Headings define the type of mutational analysis (full exome or targeted 607 gene panel), the tumour mutational burden (high/low; cut-off = 8mutations/Mb), the HPV clade (7, 9, other), the histological type (squamous, adenocarcinoma, other), first treatment received (surgery, chemoradiation, NACT), node status (positive/negative), FIGO stage (2014) not taking into account lymph node involvement and regrouped into I-II versus III-IV. Quantile PFS ordered by time interval from start to date of progression: 1. PD prior to 6 Mo, 2. 6–12 Mo, 3. 12–18 Mo, 4. DFS at last follow up.