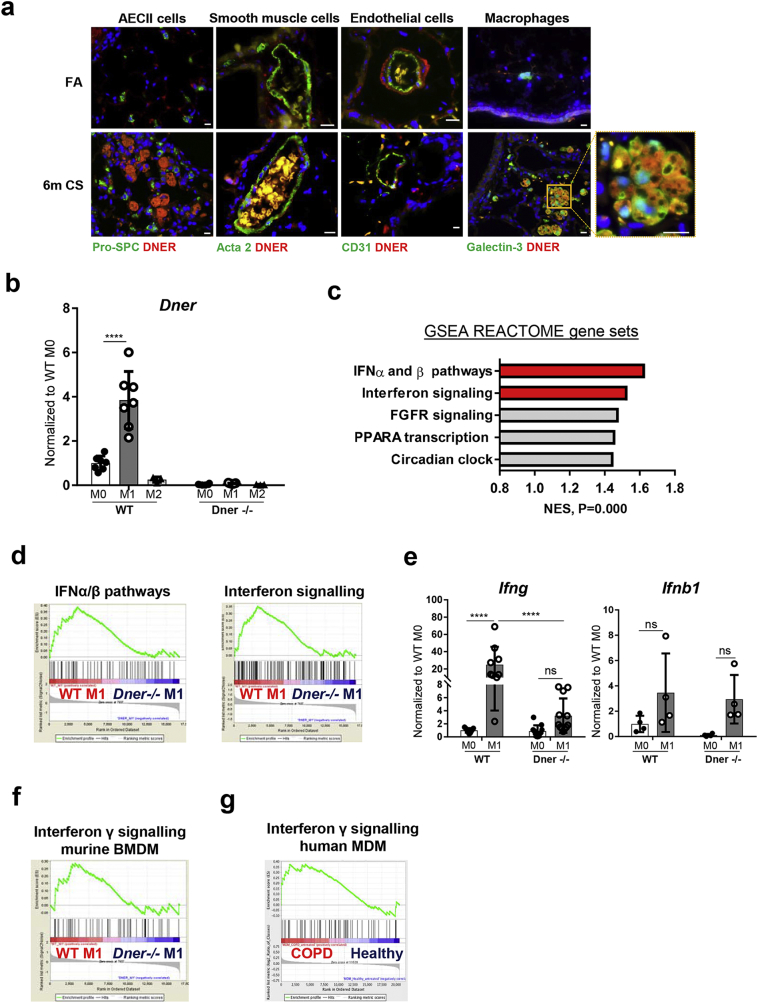
Fig. 2.
DNER is localised to macrophages in 6 m CS-exposed mice and regulates IFNγ response in M1 BMDM. (a) Representative immmunofluorescence images of lung tissue from filtered air (FA) and 6 month CS-exposed mice stained with DNER (Red), DAPI (Blue) and Galectin-3/Pro-SPC/Acta-2/CD31 (green) (n = 2–3 mice per group). Scale bar, 10 μm. (b) Dner mRNA levels in WT and DNER deficient bone marrow derived macrophages (BMDM) treated with 1 μg/ml LPS and 20 ng/ml IFNγ (M1) or 20 ng/ml IL-4 (M2) or untreated (M0). (c) Top 5 pathways with the highest Normalized Enriched Scores (NES) in Gene set enrichment analysis (GSEA) of the microarray data from WT M1 vs DNER deficient M1 BMDM using a list of 270 REACTOME pathway gene sets obtained from the Broad Institute software. (d) Enrichment plots from (2c) of WT M1 vs DNER deficient M1 BMDM. (e) mRNA levels of Ifnb1 and Ifng in M0 and M1 BMDM populations. (f) Enrichment plot of IFNγ signalling following GSEA analysis of the microarray data from WT M1 vs DNER deficient M1 BMDM. (g) Enrichment plot of IFNγ signalling from the GSEA analysis of Monocyte Derived Macrophages (MDM) obtained from healthy and COPD patient microarray data (NCBI GEO dataset, GSE8608). Two-way ANOVA, Tukey's multiple comparisons test, ****P < 0.0001 (b, e). Representative 2 independent experiments out of 5, n = 3 mice, 2–3 replicates per mouse (b, e). One experiment, n = 3 (c-d, f-g). Data shown mean values ± SD. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)