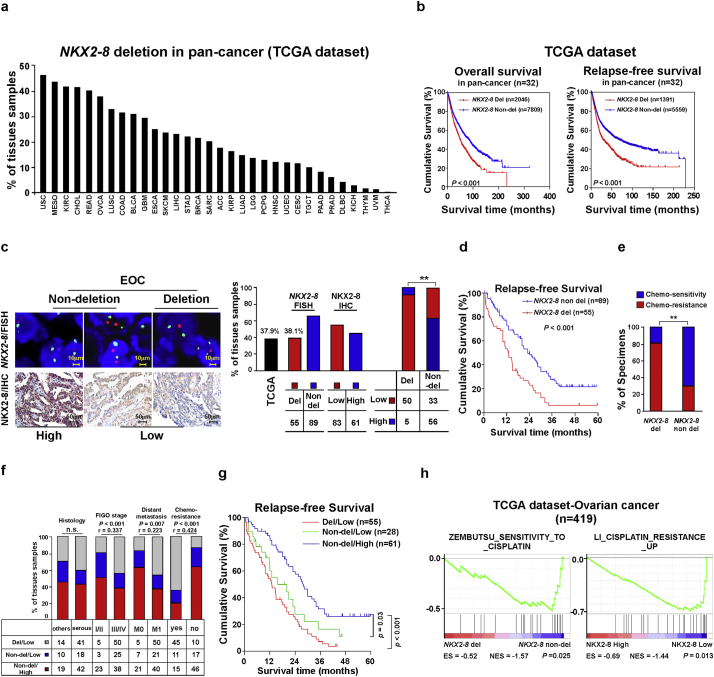
Fig. 1.
NKX2–8 deletion correlates with worse prognosis in patients with EOC. a. GISTIC2.0 analysis of 32 human cancer types (The Cancer Genome Atlas, TCGA) showing varying degrees of heterozygous deletions of NKX2–8. b. Loss of NKX2–8 was correlated with overall and relapse-free survival in the TCGA pan-cancer cohort (P < .001; n = 9822; 32 cancer types). c. EOC was analyzed by FISH for NKX2–8 deletion (left, upper), IHC for NKX2–8 protein (left, lower), and the correlation between NKX2–8 deletion and protein expression (right) (P < .01). d. Loss of NKX2–8 was associated with shorter relapse-free survival in EOC (P < .001; n = 144). e. Significant correlation between NKX2–8 deletion and chemoresistance in EOC (P < .01; n = 144). f. Significant correlation between NKX2–8 protein expression and NKX2–8 deletion with the EOC clinical pathological parameters. NKX2–8 expression was stratified into three groups: (i) NKX2–8 del/low, (ii) NKX2–8 non-del/low and (iii) NKX2–8 non-del/high. g. Kaplan-Meier survival analysis of low protein expression/NKX2–8 deletion, low protein expression/NKX2–8 non-deletion, and high expression/NKX2–8 non-deletion. h. GSEA analysis showing that NKX2–8 deletion and NKX2–8 mRNA levels was correlated with CDDP resistance-related gene signature in the TCGA ovarian cancer dataset (P < .05; n = 419).