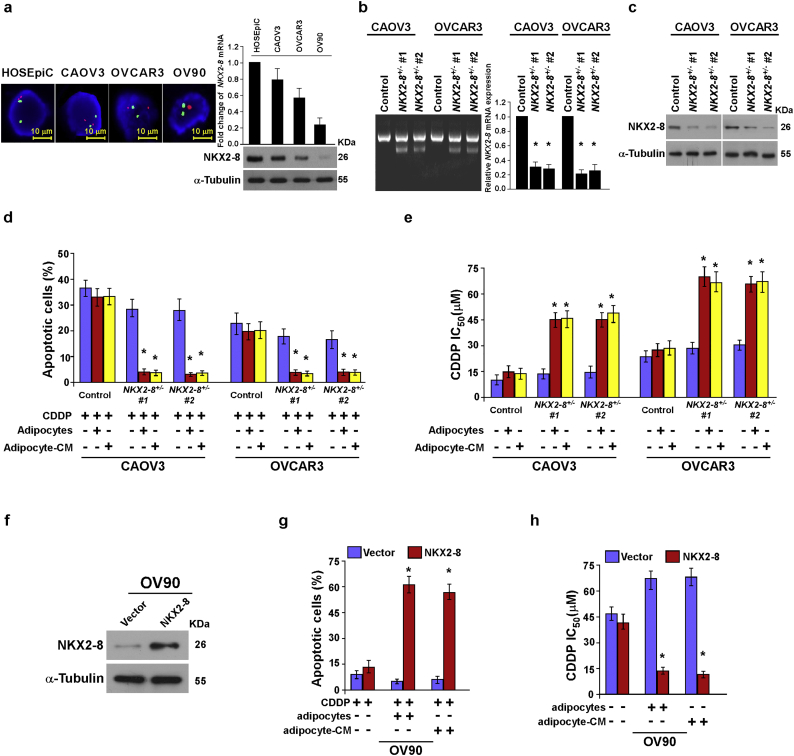
Supplementary Fig. S2.
Adipose microenvironment enhances chemoresistance of NKX2-8-deleted EOC cells. a. Ovarian cancer cell lines were analyzed using FISH with green and red probes detecting NKX2-8 locus and the chromosome 14 centromere, respectively (left). The fold change in NKX2-8 mRNA and protein expression was normalized to human ovarian surface epithelial cells HOSEpiC using quantitative RT-PCR (upper) and immunoblotting (lower), respectively. GAPDH served as a loading control. α-Tubulin served as a loading control. b. NKX2-8 heterozygous knockout cells were established using a CRISPR/Cas9 system. T7 endonuclease I cleavage confirmed the efficiency of sgRNA(left). The mRNA expression of NKX2-8 in NKX2-8 heterozygous knockout cells was detected by Real time PCR assay(right). c. NKX2-8 KO efficiency was detected by Western blotting, α-Tubulin served as a loading control. d. The apoptotic index in the indicated adipocyte-CM-treated EOC cells, as analyzed using Annexin V assay. e. Relative IC50 value for CDDP in the indicated cells treated with vehicle or Perhexiline (10 μM) and then analyzed using MTT cell viability assays in the presence of adipocyte-CM. f. Immunoblotting analysis of NKX2-8 expression in the indicated cells. g. The apoptotic index in the indicated adipocyte-CM-treated EOC cells, as analyzed using annexin V assay. h. Relative IC50 value for CDDP in the indicated cells treated with vehicle or Perhexiline (10 μM) and then analyzed using MTT cell viability assays in the presence of adipocyte-CM. Error bars in Supplementary Fig. 2d, e, g, h represent the mean ± SD of three independent experiments. ⁎P < 0.05, ⁎⁎P < 0.01.