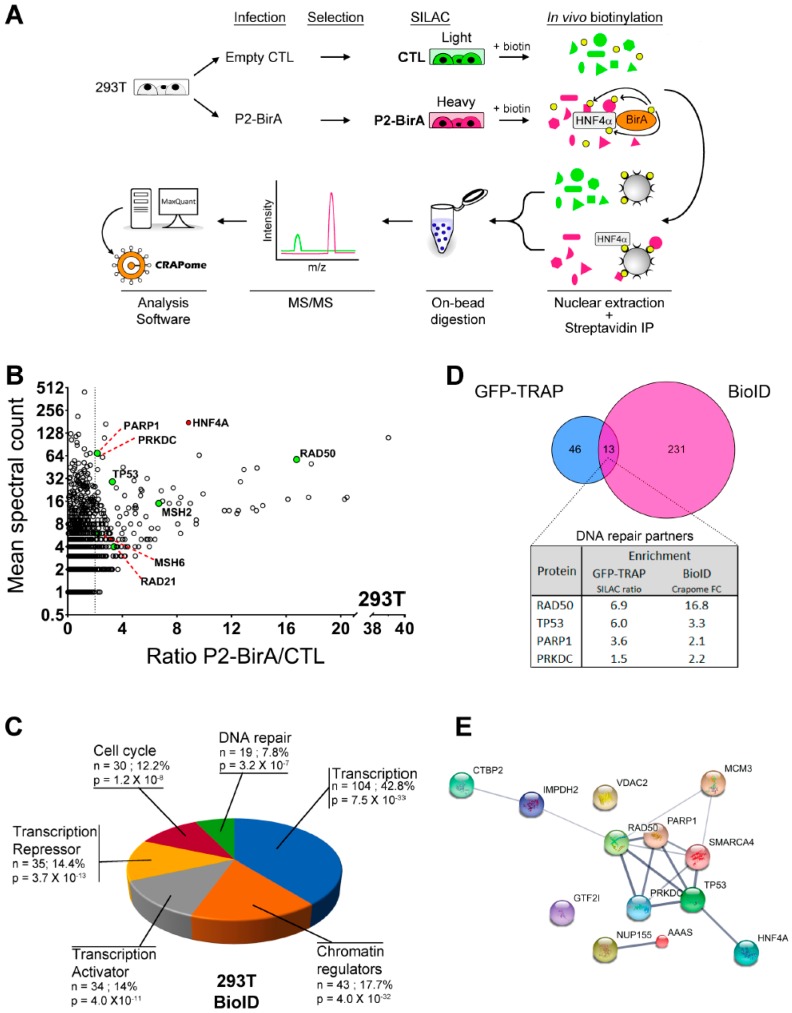
Figure 3.
BioID analysis confirms interaction of P2-BirA with DNA damage proteins in HEK293T cells. (A) Experimental design of the BioID technique used to label in vivo interacting partners of P2-BirA. HEK293T cells were infected using lentivirus expressing a construct of HNF4α tagged in C-terminal with the biotin ligase enzyme BirA (P2-BirA) or a control empty vector. After selection, the whole cell proteome was labelled by SILAC using light molecular weight isotopes for control and heavy molecular weight isotopes for P2-BirA cells. Cells were then incubated for 24 h in the presence of 50 µM of biotin allowing endogenous biotinylation of proteins surrounding the P2-BirA recombinant protein. Nuclear proteins were then extracted, and biotinylated proteins were purified using the affinity capture by streptavidin-coupled beads. The beads from the two conditions were then combined to fragment proteins in smaller peptides by on-bead digestion using trypsin. Peptides were analyzed by MS/MS and proteins were identified according to the molecular signature from the isotope labelling using the MaxQuant software and enrichment above background calculated by the CRAPome database; (B) distribution of the proteins biotinylated by P2-BirA in the HEK293T cells. The enrichment ratio of each protein in the P2-BirA condition was plotted against its mean spectral count. The proteins that are implicated in the DNA damage response have been highlighted; (C) comparison of the partners of P2-HNF4α identified by GFP-Trap or BioID. Among the 13 common targets identified, the DNA repair proteins TP53, RAD50, PARP1 and PRKDC (DNA-PKcs) were found; (D) biological function analyses by the software DAVID of the proteins biotinylated by P2-BirA in HEK293T cells. n = number of proteins identified that are associated to that function, % = percentage of the protein pulldowns with P2-GFP that are associated with that function, p = Benjamini corrected p-value; (E) interaction relationship between the 13 common targets identified by GFP-Trap and BioID based on the STRING database. Line thickness indicates the strength of the predicted relationship between the proteins.