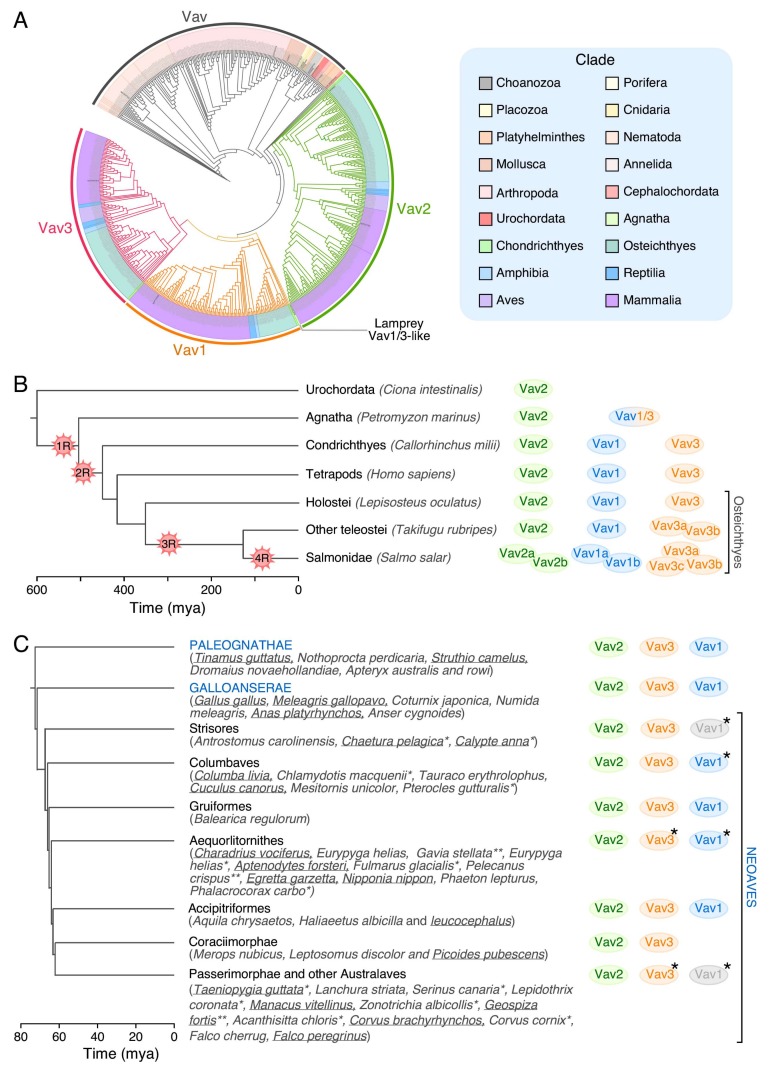
Figure 5.
The gain and loss of Vav family members in vertebrates. (A) Phylogenetic tree of Vav family proteins (left). Proteins containing DH and SH2 domains were obtained from the UniProt database and the phylogenetic tree reconstructed using the maximum-likelihood method. Node colors indicate if the protein belongs to the single Vav family group (gray color) or the vertebrate Vav1 (orange color), Vav2 (green color) or Vav3 (purple color) branch. The lamprey (Petromyzon marinus) Vav1/3-like protein node has been highlighted in light brown. The color code used for each phylogenetic clade is shown on the light blue box (right). (B) Number of Vav family genes conserved in each vertebrate clade. 1R and 2R indicate the two rounds of whole-genome duplications that took place before the divergence of jawless and cartilaginous fish, respectively. 3R and 4R indicate the lineage-specific whole-genome duplication events associated with the origin of the ancestor of most teleost fish and salmonids, respectively. Time scale is indicated at the bottom (Mya; million years ago). (C) Variability in the number of Vav family proteins in avian clades. Species with either high-coverage or fully sequenced genomes are underlined. Cases associated with the loss of Vav1 and/or Vav3 genes in some species are indicated with asterisks. Proteins colored in gray indicate that the Vav family member has been lost in a high percentage of species of the indicated group. Avian phylogeny has been established as described elsewhere [105]. Time scale is indicated at the bottom.