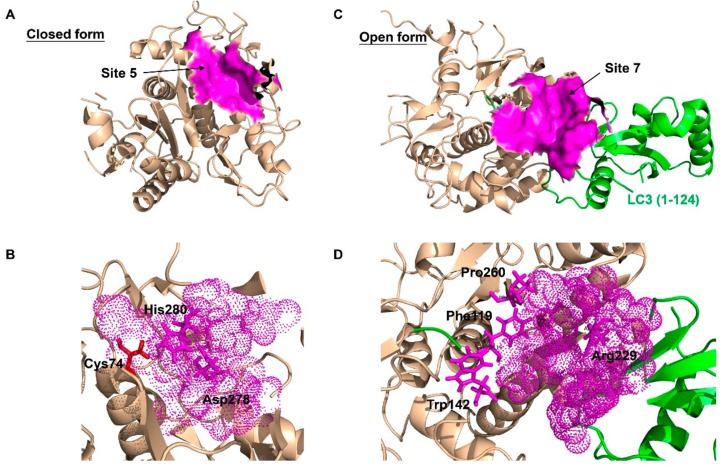
Figure 3.
The best predicted-docking pockets of the closed form and open form of ATG4B structure. (A) Crystal structure of the closed form of ATG4B protein (PDB ID: 2CY7) with labeled pocket site 5. Site 5 contains Thr10, Leu11, Ala14, Asn261, Ser262, His264, Tyr276, Asp278, His280 and Cys306. Site 5 is shown with surface and colored magenta. (B) Close-up view of site 5, where site 5 is shown with dots and colored magenta. The active site amino acids Cys74 (red), Asp278 (magenta) and His280 (magenta) are shown with sticks. Asp278 and His280 are together within the pocket surface that is suitable for molecular docking. (C) Crystal structure of the open form of ATG4B (PDB ID: 2Z0E) with labeled pocket site 7. Site 7 contains Trp142, Arg229, Leu232, Thr233, Pro260, Asp314, Pro315 and Ser316 of ATG4B, and Tyr38, Gly40, Glu41, Lys42, Gln43, Ala114, Ser115, Gln116, Glu117, Thr118 and Phe119 of LC3B. Site 7 is shown with surface and colored magenta. (D) Close-up view of site 7, where site 7 is shown with dots and colored magenta. Residues Trp142, Arg229 and Pro260 of ATG4B, and Phe119 of LC3B, which are clustered within the pocket surface and related to the interaction, are shown with sticks. The best docking pockets were defined by MOE2010 and all structure models were generated using PyMOL.