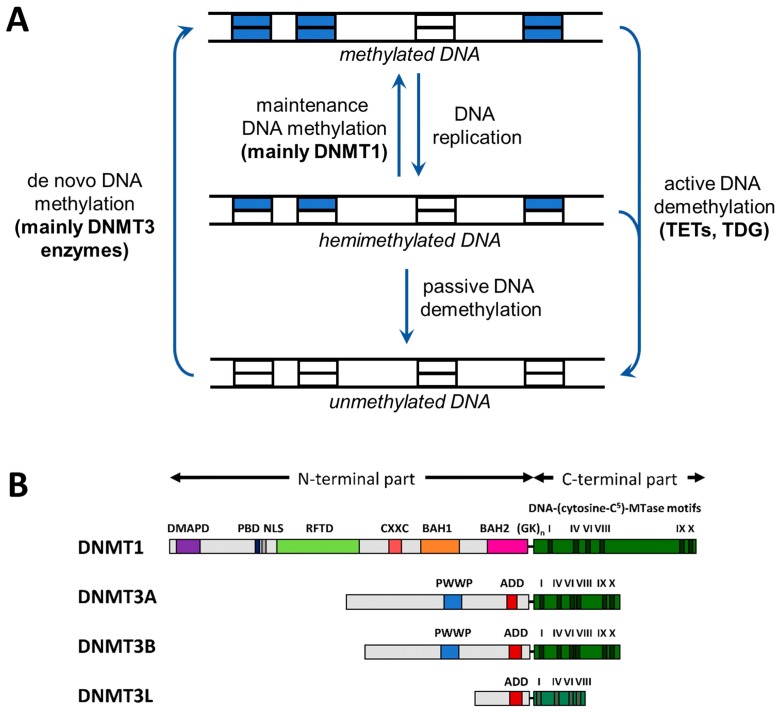
Figure 1.
Cycle of DNA methylation and domain structure of DNA methyltransferases (DNMTs). (A) Cycle of DNA methylation in human cells (adapted from [9]). DNA methylation patterns are generated by de novo methyltransferases and kept through DNA replication by maintenance methylation. DNA methylation can be lost through passive or active demethylation (abbreviations: TET, Ten-eleven translocation enzyme; TDG, thymine-DNA glycosylase). (B) Domain structure of the mammalian DNMTs DNMT1, DNMT3A, and DNMT3B. DNMT3L is a catalytically-inactive member of the DNMT3 family, which has regulatory roles [15]. The human DNMT1, DNMT3A, DNMT3B, and DNMT3L proteins consist of 1616, 912, 853, and 387 amino acid residues, respectively. Abbreviations used: DMAPD, DNA methyltransferase-associated protein 1 interacting domain; PBD, PCNA binding domain; NLS, nuclear localization signal; RFTD, replication foci targeting domain; CXXC, CXXC domain; BAH1 and BAH2, bromo-adjacent homology domains 1 and 2; GKn, glycine lysine repeats; PWWP, PWWP domain; and ADD, ATRX-DNMT3-DNMT3L domain (reprinted from [15] with permission).