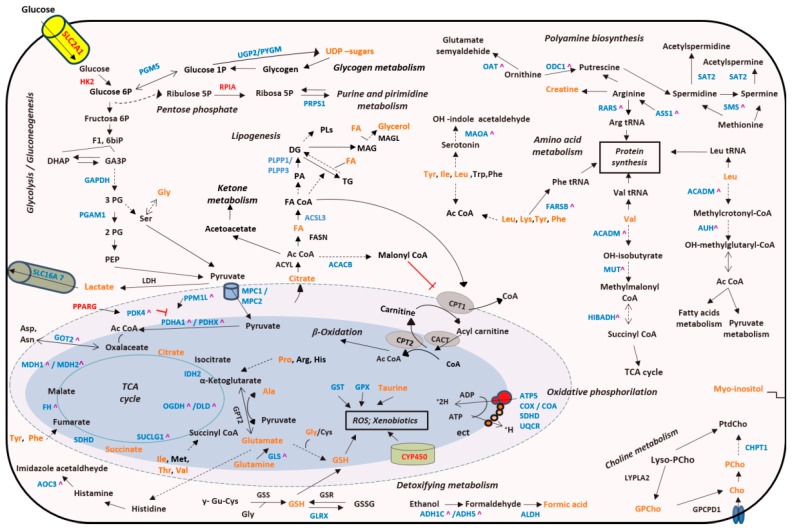
Figure 5.
Integrated altered metabolic pathways in bladder tumors according to our metabolomic and transcriptomic data. Note: Enzymes shown in red indicate significant overexpression in BC tissue; enzymes shown in blue indicate significant downexpression; enzymes shown in black indicate no significant changes in our studies; metabolites shown in orange indicate that they were identified in our studies; ^ Indicate that these genes presented a differential alternative splicing in our tumor tissue samples; Dashed lines indicate that the reaction is not direct. Ac CoA: acetyl coenzyme A; ADP: adenosine diphosphate; Ala: alanine; Arg: arginine; Asn: asparagine; Asp: aspartate; ATP: adenosine triphosphate; 1;3 biPG: 1,3-bisphosphoglycerate; Cho: choline; CoA: coenzyme A; DG: diacylglycerides; DHAP: dihydroxyacetone phosphate; FA: fatty acids; F1,6biP: fructose-1,6-bisphosphate; GA3P: glyceraldehyde 3-phosphate; Glu: glutamate; Gly: glycine; GPCho: glycerophosphocholine; GSH: glutathione; His: histidine; Ile: isoleucine; Leu: leucine; Lys: lysine; MAG: monoacylglycerols; NO: nitric oxide; PA: phosphatidic acid; PEP: phosphoenolpyruvate; 2PG: 2-bisphosphoglycerate; 3PG: 3-bisphosphoglycerate; Phe: phenylalanine; PCho: phosphocholine; PLs: phospholipids; Pro: proline; ROS: reactive oxygen species; Ser: serine; TCA cycle: tricarboxylic acid cycle; TG: triacylglycerides; Trp: tryptophan; Tyr: tyrosine; UDP-sugars: uridine diphosphate-sugars; tRNA: transfer ribonucleic acid. The names of the significant metabolic genes are detailed in the Supplementary Table S4.