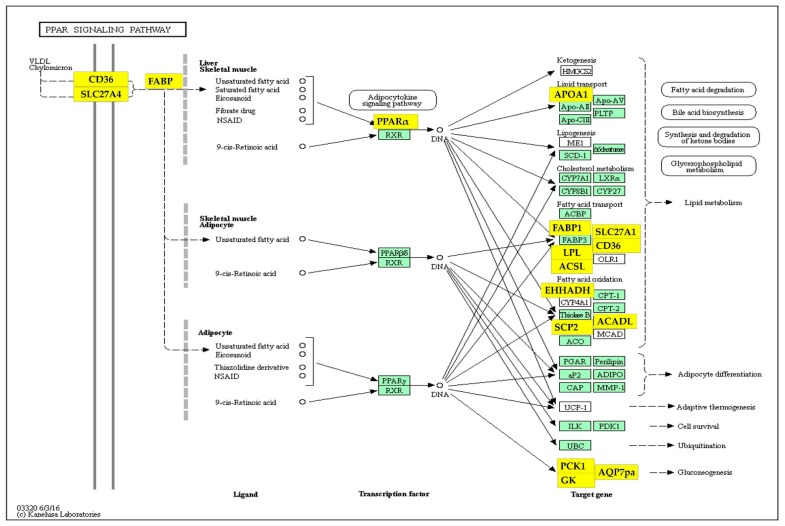
Figure 2.
PPAR (peroxisome proliferator-activated receptor) signaling pathway enriched of differentially expressed genes (DEGs) in the jejunum of broiler chickens. The PPAR signaling pathway as the DAVID (Database for Annotation, Visualization and Integrated Discovery) functional annotation significantly enriched DEG is visualized by the Kyoto Encyclopedia of Genes and Genomes (KEGG) [10] pathway mapper. Genes over-expressed in the jejunum are shown in a yellow color. CD36: CD36 molecule; ACADL: acyl-CoA dehydrogenase long chain; ACSL: acyl-CoA synthetase long-chain family member (ACSL3, ACSL4, ACSL5); APOA1: apolipoprotein A-1; AQP7: aquaporin 7; EHHADH: bi-enzyme enoyl-CoA hydratase/3-hydroxy acyl CoA dehydrogenase; FABP: fatty-acid binding protein (FABP1, FABP2, FABP5, FABP6); GK: glycerol kinase; LPL: lipoprotein lipase; PPARα: peroxisome proliferator-activated receptor alpha; PCK1: phosphoenolpyruvate carboxykinase 1; SCL27A1/4: solute carrier family 27 (fatty-acid transporter), member 1, member 4; SCP2: solute carrier protein 2.