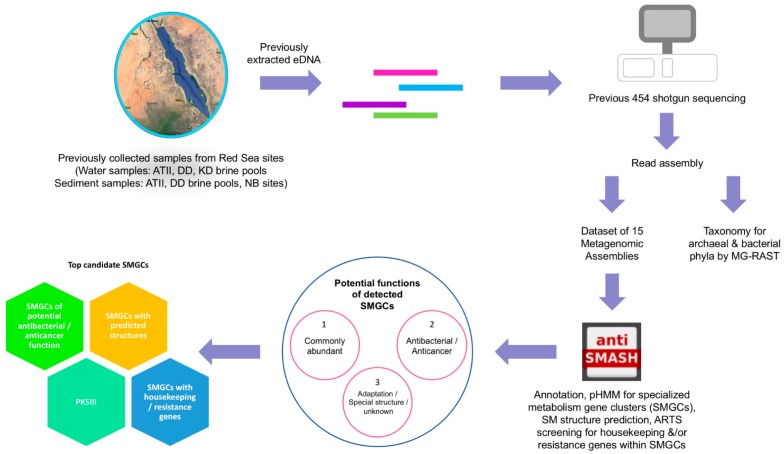
Figure 1.
Study workflow for the analysis of specialized metabolism gene clusters in Red Sea brine pools. Water and sediment samples were earlier collected from Red Sea Atlantis II Deep (ATII), Discovery Deep (DD), Kebrit Deep (KD) brine pools and brine-influenced (NBI, NBII) sites. Metagenomic prokaryotic DNA was then extracted from each site and 454 shotgun sequencing was performed followed by read assembly. Taxonomic classification for archaeal and bacterial phyla was performed by protein-based phylogeny using the metagenomics rapid annotation using subsystems technology (MG-RAST) tool [37]. The metabolite analysis shell (AntiSMASH) tool was then used for annotation, for identifying specialized metabolism gene clusters (SMGCs) by translated amino acid sequence comparison with signature biosynthetic genes profile hidden Marcov Model (pHMMs), and for structure prediction of the specialized metabolites [9]. The Antibiotic Resistance Target Seeker (ARTS) tool was used to detect housekeeping and/or resistance genes within the SMGCs [38]. The predicted specialized metabolites were grouped by their potential functions into three major groups. Lastly, top candidate SMGCs were identified in the Red Sea brine dataset.