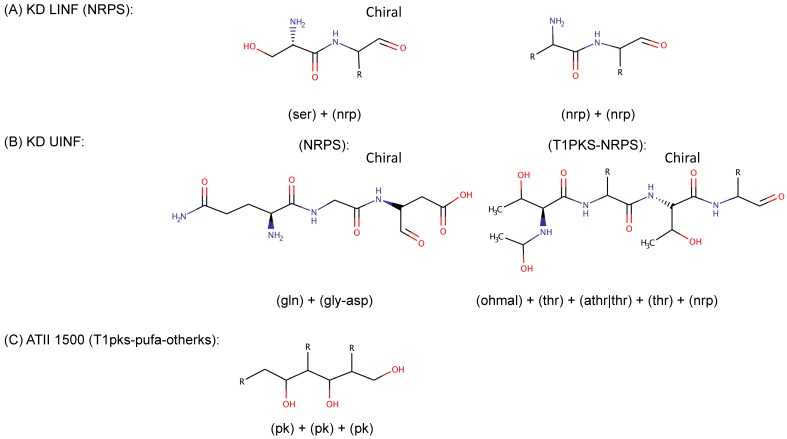
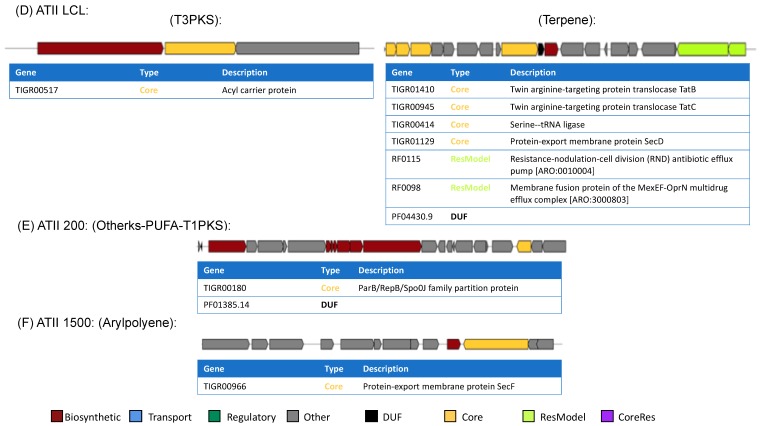
Figure 3.
Promising Red Sea brine specialized metabolite candidates. Five core structures for specialized metabolites were predicted using antiSMASH 4.0 [45]: (A) KD brine–seawater Lower Interface (KD LINF) site had two non-ribosomal peptides predicted to be synthesized by two non-ribosomal peptide synthetase (NRPS) clusters. (B) KD brine–seawater Upper Interface (KD UINF) site had a non-ribosomal peptide predicted to be synthesized by a NRPS cluster and a hybrid polyketide-non-ribosomal peptide predicted to be synthesized by T1PKS-NRPS. (C) ATII 1500 site had a polyketide predicted to be produced by a T1pks-pufa-otherks hybrid cluster. Four top promising candidate SMGCs were detected by ARTS [38]: (D) ATII brine Lower Convective Layer (ATII LCL) site had two clusters encoding Terpene and T3PKS. (E) ATII 200 site had a cluster encoding aryl polyene. (F) ATII 1500 site had a cluster encoding Otherks-PUFA-T1PKS (PUFA: polyunsaturated fatty acid). ser: serine, nrp: non-ribosomal peptide, gln: glutamine, gly: glycine, asp: aspartate, thr: threonine, pk: polyketide, ohmal: hydroxy malate.