Figure 4.
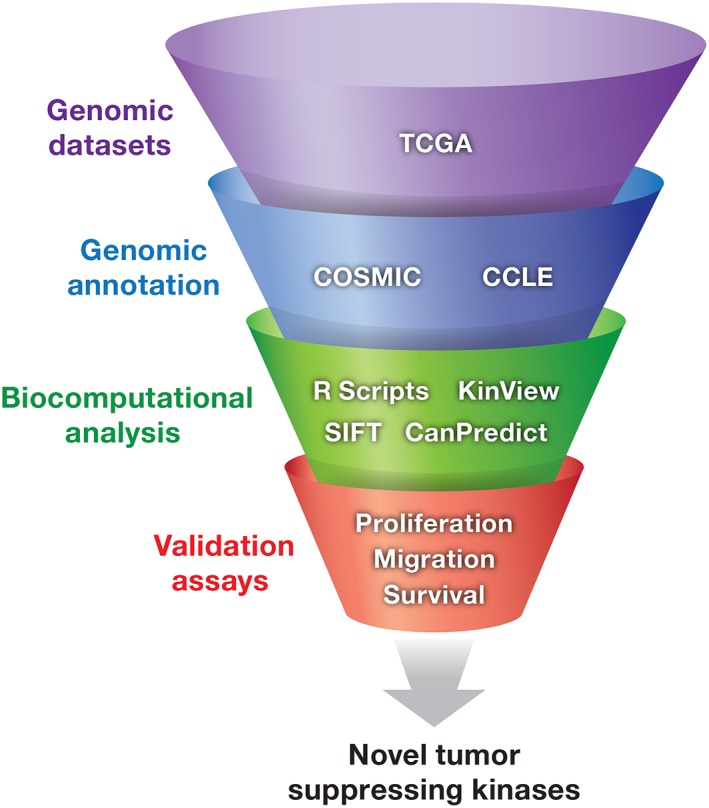
The Tumor suppressing kinome. Filtering of Cancer genomic datasets with high‐throughput mutation assessors that are biased toward predicting functional mutations in highly conserved residues or R scripts designed to identify mutations in critical residues required for catalytic activity greatly aid in identifying novel tumor suppressing kinases. Validation is then required through biochemical and functional assays.
