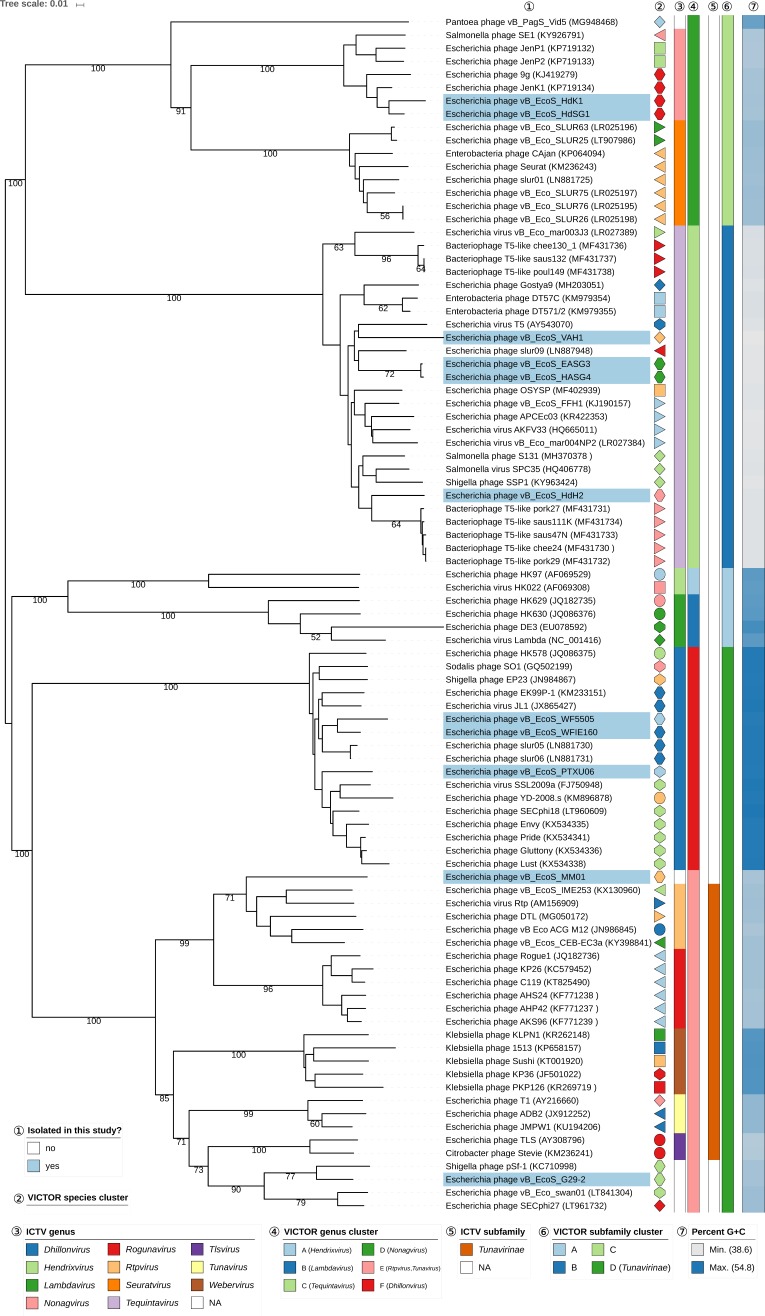
Figure 6.
Phylogenomic analysis of Siphoviridae members of this study at the amino acid level using VICTOR. The recommended VICTOR tree (formula d6) is shown and the numbers listed above the branches are GBDP pseudo-bootstrap support values from 100 replications, given that branch support exceeds 50%. The branch lengths of the resulting VICTOR trees are scaled in terms of the used distance formula. Annotations are given to the right-hand side of the tree: indicator for newly isolated phages (1), genera and subfamilies according to ICTV (3,5), species, genus, and subfamily cluster proposed by VICTOR (2,4,6), and genomic G+C content (7).