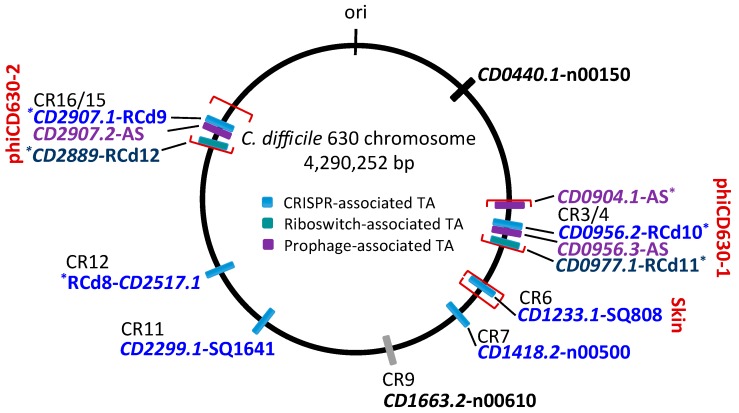
Figure 1.
Schematic genomic map of type I TA loci in C. difficile strain 630. The location of type I TA modules is shown in blue for CRISPR-associated loci, in green for c-di-GMP-responsive riboswitch-associated loci, and in purple for additional prophage-associated TA loci encoding 34–35 amino acid proteins. The prophage position is indicated using square brackets for phiCD630-1, phiCD630-2, and skin element. * indicates TA modules with detailed characterization. CRISPR arrays (CR) are numbered according to CRISPRdb (https://crispr.i2bc.paris-saclay.fr/crispr/) and previous publications [23,26,27,28]. The CD1663.2-n00610 locus associated with CRISPR 9 array but encoding a small protein with a divergent sequence is indicated in grey. “ori” indicates the origin of replication.