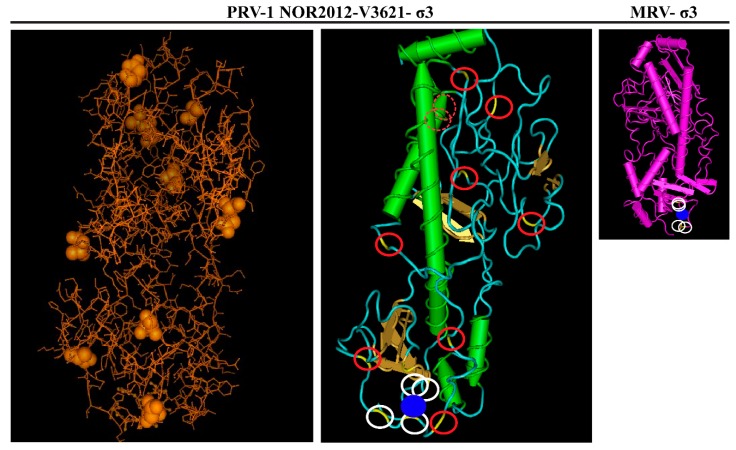
Figure 5.
Visualization of the predicted 3D-structure of the NOR-2012 σ3 protein with Vector NTI 3D molecule viewer Cn3D v4.3 following structure homology modeling using the MRVσ3 protein (PDB ID:1FN9) as template, the most appropriate structure model to use as predicted by i-TASSER. The ten amino acid sites differing between the HSMI associated (here represented by NOR-2012) and low virulent HSMI isolates are indicated by or as yellow/brown balls (left picture) or as a yellow color enclosed by a red circle (middle picture). Amino acid side chains predicted to coordinate the Zn-ion, represented by a blue ball, are indicated by white circles. MRV-σ3 added for comparison.