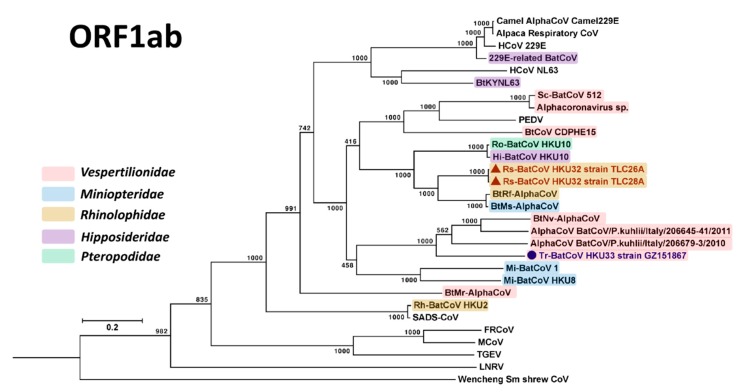
Figure 3.
Phylogenetic analysis of ORF1ab amino acid sequences of Rs-BatCoV HKU32 strains TLC26A and 28A, Tr-BatCoV HKU33 strain GZ151867 and other alphaCoVs. ORF1ab tree was constructed by maximum likelihood method using LG + G + I + F substitution model. The bootstrap values are calculated from 1000 trees. Tree was rooted using corresponding sequence of Middle East respiratory syndrome (MERS)-CoV (GenBank accession number YP_009047202.1). All bootstrap values are shown. The scale bar represents 5 substitutions per site. Both Rs-BatCoV HKU32 and Tr-BatCoV HKU33 are labeled with red (triangle) and blue (circle), respectively. Corresponding viral bat hosts’ families are highlighted in different colors: Red, Vespertilionidae; blue, Miniopteridae; yellow, Rhinolophidae; purple, Hipposideridae; green, Pteropodidae.