Figure 1.
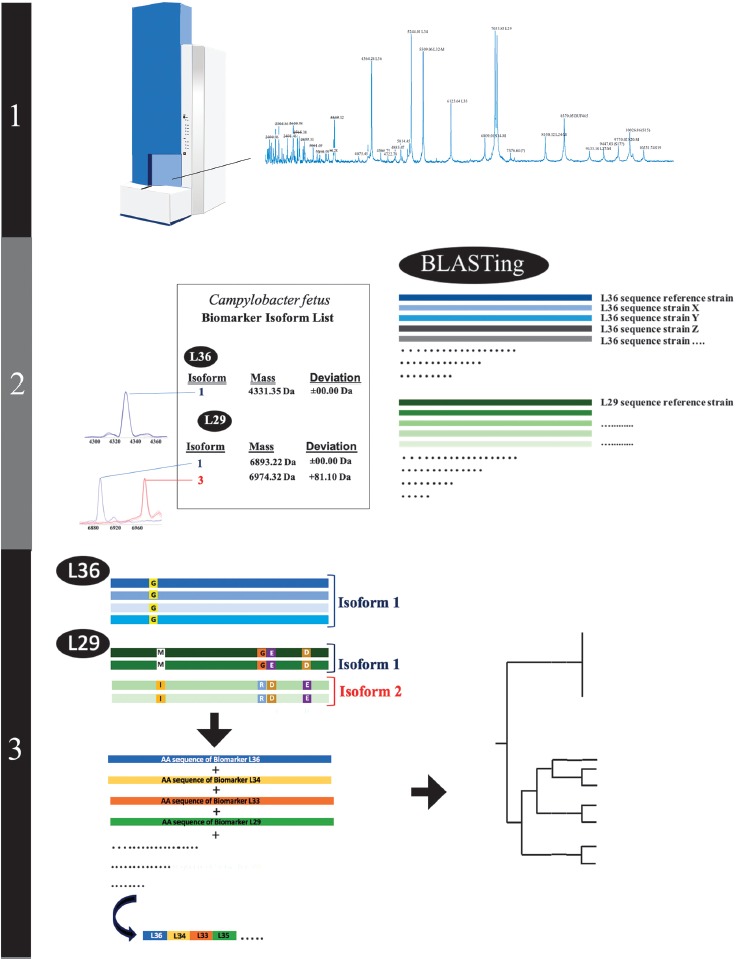
Illustration of the different proteotyping steps. 1) Recording of the ICMS mass spectra of the C. fetus test cohort and reference strain Cff LMG 6442 (NCTC 10842). 2) Establishment of a C. fetus-specific allelic isoform list by blasting the genome sequences obtained from the NCBI database against the genome of the C. fetus reference strain. Subsequently, allelic isoforms in the test cohort are identified by comparing with the newly established allelic isoform list. 3) For each strain in the test cohort, a specific set of biomarker isoforms is obtained. Subsequently the amino acid sequences of the biomarkers are fused into a single sequence that results in specific proteotyping-based sequence type for each of the strains and allows the calculation of an proteotyping-derived taxonomic dendrogram
