Figure 4.
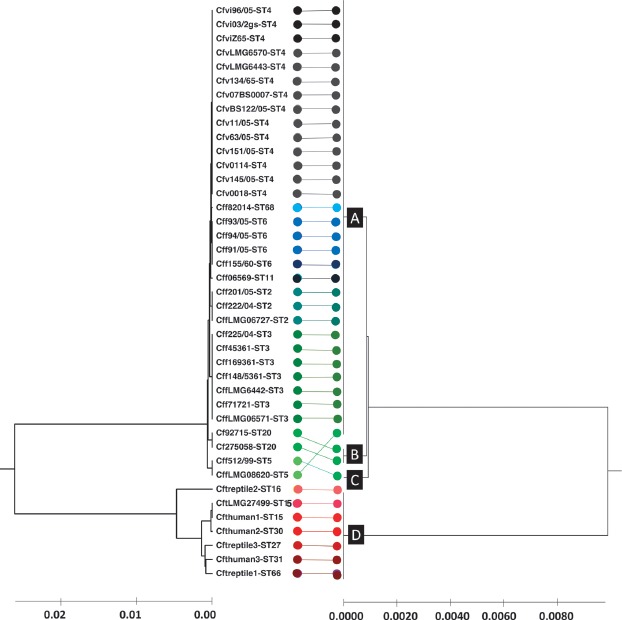
Comparison of MLST- and proteotyping-derived phylogenies. On the left: Evolutionary tree calculated based on MLST by means of the maximum composite likelihood method (UPGMA). In total, 14 different MLST sequence types were identified which are illustrated in different colors. On the right: Evolutionary tree based on proteotyping and calculated using UPGMA. Four different proteotyping-derived types were identified. Type A contains most of the C. fetus subsp. fetus and C. fetus subsp. venerealis strains. Type B and C contain 2 MLST ST 5 and one MLST ST 20 strain. The most interesting proteotyping-derived type is type D, which contains all C. fetus subsp. testudinum strains and thereby allows the differentiation of the subspecies from other C. fetus subspecies. The different proteotyping-based sequence types are marked at the branches of the evolutionary tree (A, B, C, and D).
