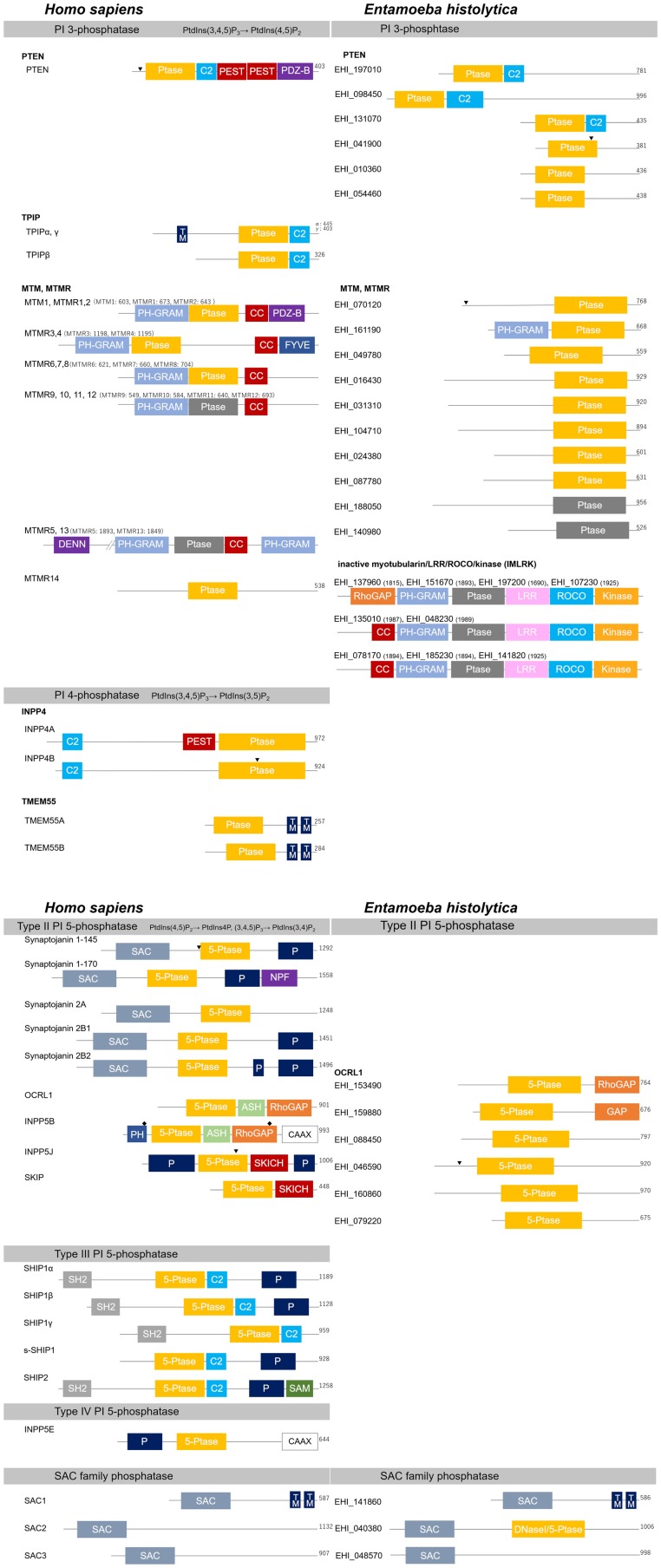
Figure 4.
Structural features of PI phosphatases of H. sapiens and E. histolytica. Structural features and domain organization of the PI phosphatases are shown. Numbers showing at the end of the protein or after the name of the protein indicates amino acid length. 5-Ptase, PI 5-phosphatase domain; ASH, ASPM-SPD2-hydin; C2, C2 domain; CAAX, CAAX motif; CC, coiled coil domain; DENN, differentially expressed in normal vs. neoplastic; DNaseI, DNase I-like domain; FYVE, Fab1, YOTB, Vac1, and EEA1 domain; GRAM, glucosyltransferases Rab-like GTPase activators and myotubularins; GAP, GTPase-activating domain; kinase, protein kinase domain; LRR, leucine-rich repeats; NPF, asparagine- proline-phenylalanine repeats; P, Proline-rich; PDZ-B, PDZ domain binding domain; PEST, Proline, glutamine, serine, threonine; PH, Pleckstrin-homology; (Ptase; gray), inactive Ptase domain; (Ptase; yellow), active CX5R motif containing PI 3- or PI 4-phosphatase domain; ROCO, comprised of a ROC (Ras of complex proteins) and COR (C-terminal of ROC) region; SAC, Sac domain; SAM, sterile alpha motif; SH2, Src homology 2; SKICH, SKIP carboxylhomology; TM, transmembrane domain. Clathrin-binding domain and the nuclear localization signal are also labeled with “♦” and “▾”, respectively.