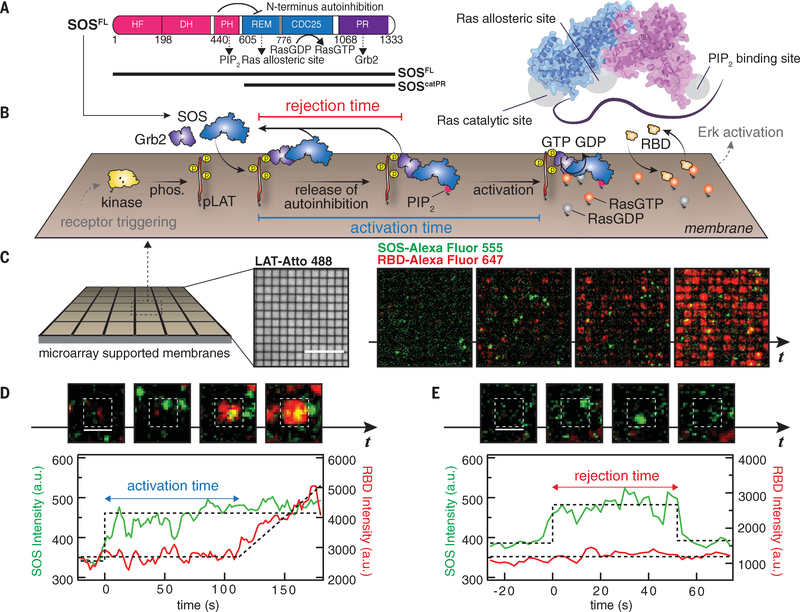
Fig. 1. Single-molecule activation assay of SOSFL on supported membranes.
(A) Domain architecture (left) and a cartoon model (right) of SOSFL The crystal structure was rendered with Protein Data Bank entry 3KSY. HF, histone fold; DH, Dbl homology domain; PH, Pleckstrin homology domain; REM, Ras exchanger motif. Numbers indicate amino acid positions. (B) Schematic of the reconstitution of Grb2-mediated SOS activation. LAT phosphorylated by membrane-bound kinase Hck (pLAT) and Ras preloaded with GDP were decorated on the supported membranes corralled by 1-μm by 1-μm or 2-μm by 2-μm chromium grids. The injection of Grb2, SOSFL-Alexa Flour 555, RBD-K65E-Alexa Fluor 647, and GTP into the solution triggered SOS recruitment, subsequent release of autoinhibition, and activation. Receptor triggering and downstream activation (in gray) are shown to illustrate the signaling pathway, and not included in the experiments. phos., phosphorylation. (C) Snapshot images of SOS (green) and RBD (red) recruitment in microarray supported membranes. Scale bar, 10 μm. t, time. (D) Definition of activation time. (E) Definition of rejection time. [(D) and (E)] Scale bars, 2 μm. a.u., arbitrary units.