FIGURE 7.
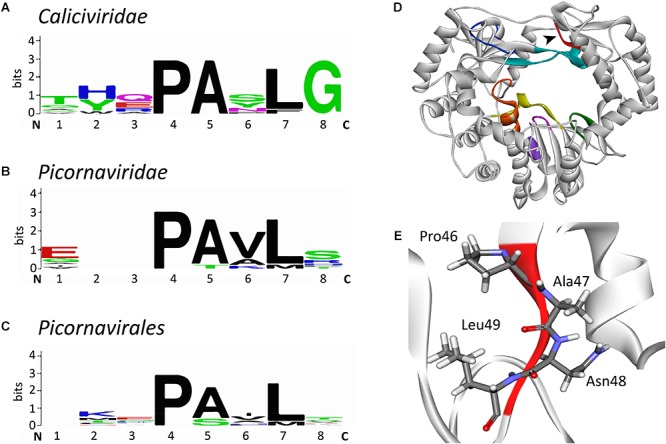
Sequence alignment logos of a putative new conserved motif (“motif I”) and the localization of the motif in the RHDV RdRp. (A) Sequence logo alignment for the putative motif of the following viruses in the family Caliciviridae: European brown hare syndrome virus and Rabbit haemorrhagic disease virus (both genus Lagovirus); Norwalk virus, Lordsdale virus, Murine norovirus (genus Norovirus); Sapporo virus (genus Sapovirus); Feline calicivirus, Vesicular exanthema of swine virus, and San Miguel sea lion virus (genus Vesivirus); Newbury 1 virus (genus Nebovirus). (B) Sequence logo alignment for the putative motif of the following viruses in the family Picornaviridae: Poliovirus, Bovine enterovirus, Coxsackievirus B3, Human rhinovirus A, and Echovirus (genus Enterovirus); Foot and mouth disease virus (genus Aphtovirus); Hepatitis A virus (genus Hepatovirus); Human parechovirus (genus Parechovirus); Theiler’s murine encephalomyelitis virus and Encephalomyocarditis virus (genus Cardiovirus); Avian encephalomyelitis virus (genus Tremovirus). (C) Sequence logo alignment for the putative motif of the following viruses in the order Picornavirales: Poliovirus, Foot and mouth disease virus, Hepatitis A virus, and Human parechovirus (family Picornaviridae); Cricket paralysis virus and Drosophila C virus (family Dicistroviridae); Parsnip yellow fleck virus, Broad bean wilt virus, Cowpea mosaic virus, and Beet ringspot virus (family Secoviridae). N and C indicate N- and C-terminal directions, respectively. Sequence conservation is measured in bits and is indicated by the height of each letter’s stack. Amino acids are colored according to their chemical properties: polar amino acids (Gly, Ser, Thr, Tyr, Cys, Gln, Asn), green; basic (Lys, Arg, His), blue; acidic (Asp, Glu), red; and hydrophobic (Ala, Val, Leu, Ile, Pro, Trp, Phe, Met), black (Crooks et al., 2004). (D) Ribbon diagram of Rabbit haemorrhagic disease virus RdRp (PDB ID: 1KHW). The black arrow head points at the new motif I that is colored red, other conserved motifs are colored as in Figure 3D. (E) Structure of the motif I. Sequence alignments were performed with the multiple sequence alignment tool MUSCLE (Edgar, 2004); sequence logo pictures were created with Weblogo (Crooks et al., 2004). The ribbon diagram was generated using Discovery Studio (Dassault Systèmes BIOVIA, Discovery Studio Visualizer v17.2.0).
