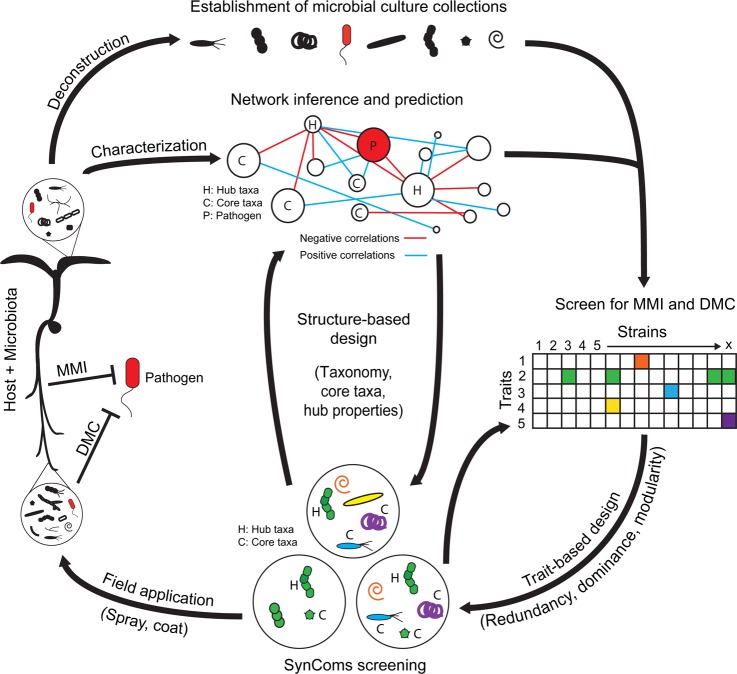
Fig 2. Rational design of SynComs with predictable pathogen biocontrol activities.
Starting in the field where a pathogen outbreak has occurred, the microbiota of diseased and healthy plants are characterized and isolated. The isolates are screened in binary microbe–microbe and in planta ternary interactions to detect and catalogue traits linked to DMC and MMI. From the obtained catalogue, activities of individual strains can be used to design more complex SynComs while taking into account trait redundancy, dominance, and modularity. In parallel, interaction networks are inferred from sequencing data, and potential key organisms are identified based on hub structural properties or functional modules. This network inference helps with prioritizing candidate strains for targeted screening of DMC and MMI traits. Both trait-based and structure-based approaches can inform the rational design of SynComs with stable and effective biocontrol activities in the field. DMC, direct microbial competition; MMI, microbiota-modulated immunity; SynComs, synthetic microbial communities.