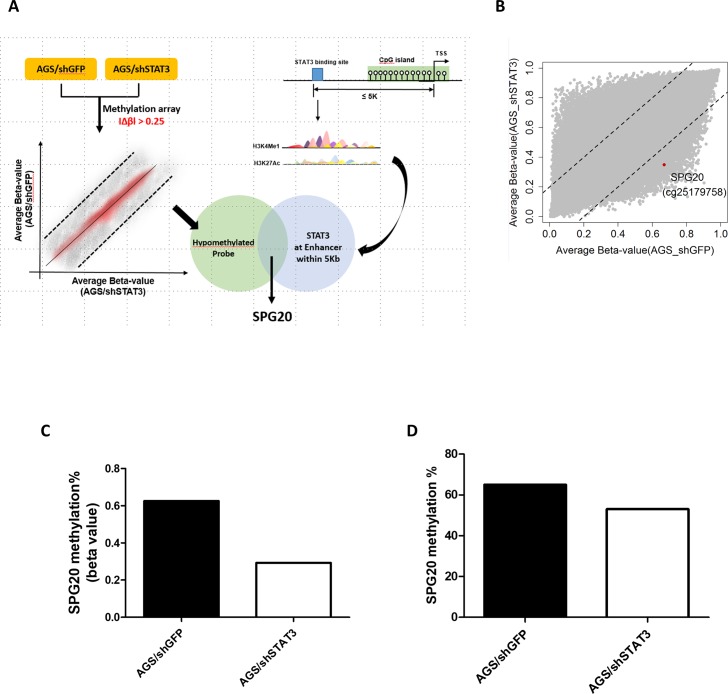
Fig 1. Integrated microarray and bioinformatics analyses identifies SPG20 as an epigenetically silenced target of STAT3, in gastric cancer.
(A) Schematic diagram showing the experimental scheme of this study. Stable transfectants of AGS gastric cancer cells depleted of STAT3 (shSTAT3) or control (shGFP) were subjected to Illumina 850K methylation microarray analysis. Probes showing a changes in β values (|Δβ|) of at least 0.25 (25% methylation difference) were selected. In addition, bioinformatics analysis was performed to identify potential STAT3 targets by filtering genes with at least one STAT3-binding site in potential enhancer region (as enriched with H3K4me1 and H3K27Ac) of the promoter CpG island. One hypomethylated probe and a potential STAT3 target, SPG20, was selected for further analysis. (B) Scatter plot showing the average beta-value of each probe in AGS cells depleted of STAT3 or GFP (as control). One of the probes (cg25179758, red dot) showing hypomethylation, located within the SPG20 promoter region, was selected for further analysis. Methylation% of SPG20 at the promoter region as determined by (C) methylation microarray (beta value) or (D) bisulphite pyrosequencing.