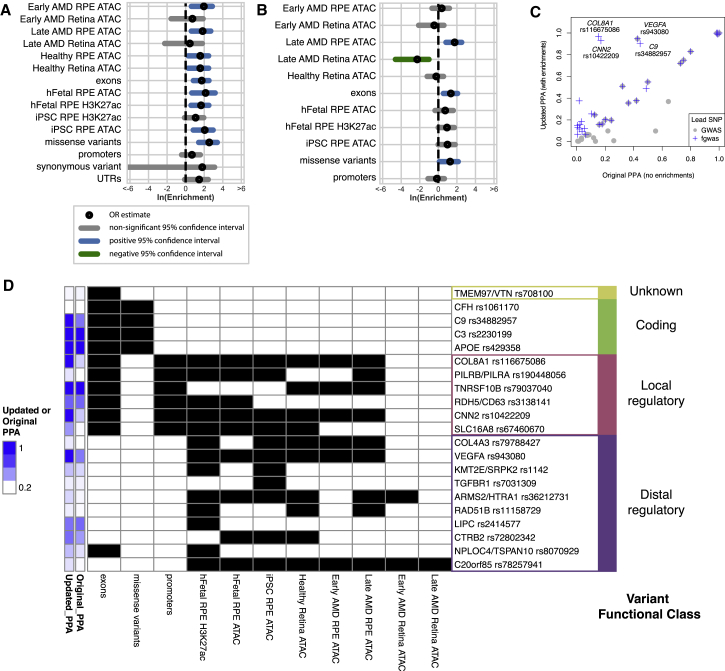
Figure 3.
Fine Mapping of AMD GWAS Loci
(A) fgwas single enrichment of GWAS signal for multiple genome annotations. Black circles indicate ln(OR) and blue or gray lines show 95% CIs. Blue indicates a significant positive enrichment.
(B) Final combined model used by fgwas to prioritize variants. Black circles indicate ln(OR) and blue, gray, or green lines show 95% confidence intervals (colored to indicate significance different from 0; green negative, blue positive). Note that in this combined model, associations reflect independent associations for each annotation and exclude annotations that did not significantly improve the model, and so differ from single enrichments shown in (A).
(C) Scatterplot showing posterior probability of association (updated PPA) versus original posterior probability of association (original PPA) for GWAS loci. GWAS lead SNPs are shown in gray. SNPs that were chosen as variant at the locus with the highest PPA by fgwas are shown in blue. When the variant with the highest updated PPA is the GWAS lead variant, the points overlap. The four variants where the fgwas prediction improved the PPA from <0.5 to >0.8 are labeled by their candidate gene and fgwas lead SNP.
(D) Heatmap indicating the presence (black) or absence (white) of each annotation for the 21 lead fgwas variants that were prioritized by the model. The original and updated PPAs are shown in blue and are the same as in (C). Variants are grouped into variant functional classes according to whether they were unknown, coding, local regulatory (located in promoters), or contained in distal regulatory (ATAC-seq or H3K27ac without promoter) annotations.
See also Figure S2.